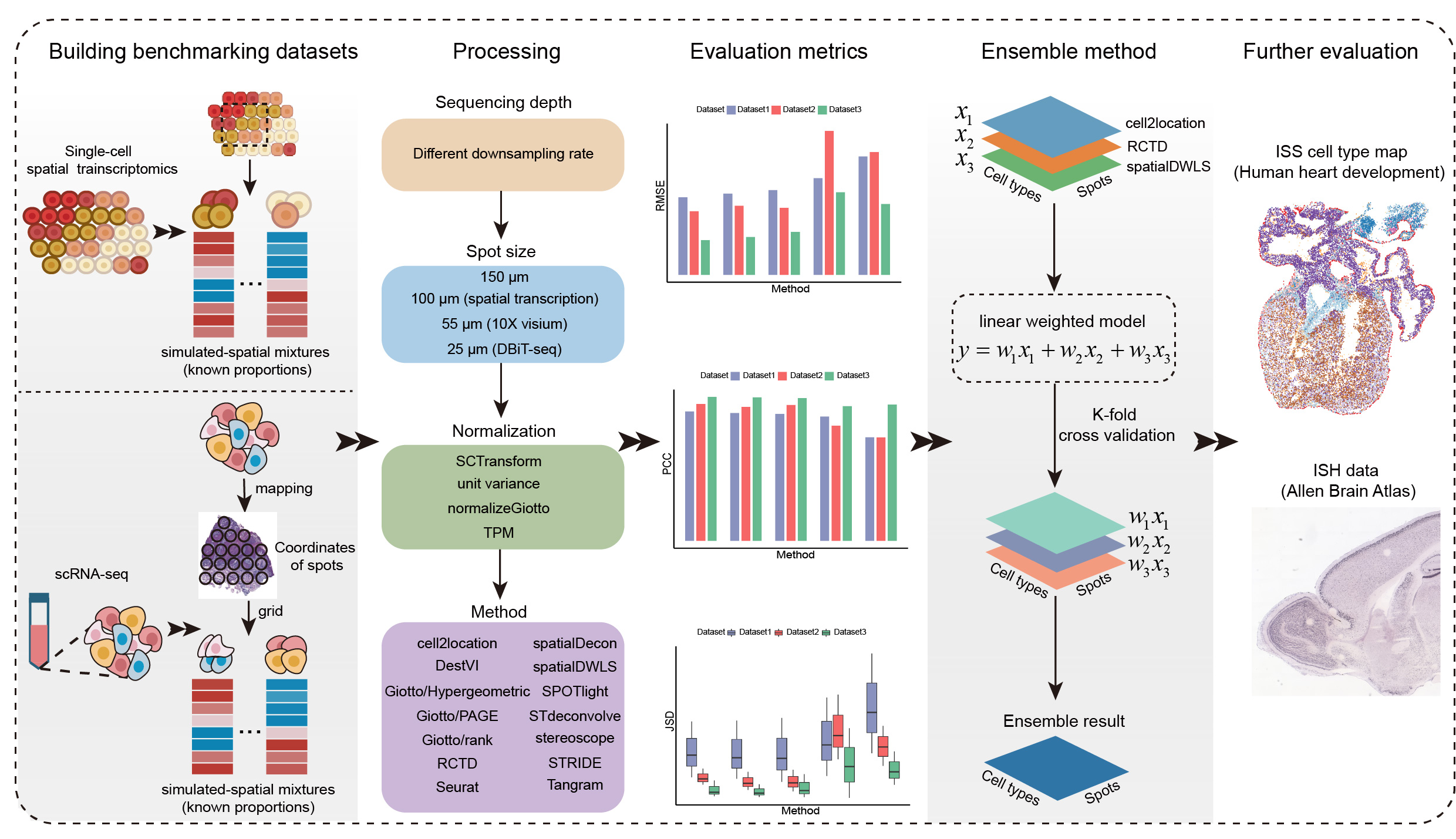
We collect three synthetic ST datasets with known single-cell compositions and a human heart ST dataset with known regional cell type information to benchmark 14 different deconvlution methods. Furthermore, we investigate the robustness of different methods to sequencing depth, spot size, and the choice of normalization. Moreover, we propose a new ensemble learning-based deconvolution method (EnDecon) by integrating multiple individual methods for more accurate deconvolution. The main steps of benchamrking pipeline include:
- We employ two staregies to generate synthetic ST datasets with known cell type compositions as ground truth. For single-cell resolution ST data, we adopted 'grid' method to simulated ST data. For scRNA-seq data, we first map it to the spatial locations of ST data to synthesize single-cell resolution ST data, and then simulate spatial mixtures with known cell type proportions using the ‘grid’ method.
- We investigate the robustness of different methods to sequencing depth, spot size, and the choice of normalization.
- We use RMSE, PCC, JSD to evaluate the performance of the 14 computational methods in disentangling cell type contribution from a single capture spot based on the above synthetic ST datasets.
- We develope an ensemble learning-based deconvolution method (EnDecon) for ST data by drawing on strengths from existing methods.
- We adopt in situ sequencing (ISS) data and in situ hybridization (ISH) data to further evaluate the performance of all methods
The respository mainly consists of the following modules:
Evaluationcontains the script to calculate the RMSE, PCC, JSDST_Deconvolutioncontains the scripts to reproduce 14 deconvolution methods and the proporsed EnDecon methodsynthetic_st_datasetcontains the scripts to generate the synthetic ST datasetsbenchamarking.Rcontains the script to benchmark different deconvolution methods on your own datasets.
Note that the cell2location, DestVI, stereoscope, STRIDE, and Tangram are written in Python and each requires different Python versions, so benchmarking.R does not include these Python-based tools.
All datasets can be downloaded from their respective sources:
- mouse embryo was downloaded from the National Center for Biotechnology Information (NCBI) under GSE166692
- MPOA was downloaded from https://datadryad.org/stash/dataset/doi:10.5061/dryad.8t8s248/
- mouse brain ST data was download from https://www.dropbox.com/s/azjysbt7lbpmbew/brain_st_cortex.rds?dl=0
- mouse brain scRNA-seq data was downloaded from https://www.dropbox.com/s/ruseq3necn176c7/brain_sc.rds?dl=0
- human developing heart ST data was downloaded from https://data.mendeley.com/datasets/mbvhhf8m62/2
- human developing heart ISS data was obtained from https://doi.org/10.6084/m9.figshare.10058048.v1
- mouse brain ISH image data was downloaded from https://mouse.brain-map.org/.
R
- RCTD (by spacexr of version 2.0.0)
- Giotto/PAGE/RANK/Hypergeometric (Version 1.1.0)
- spatialDWLS (by Giotto of Version 1.1.0)
- SPOTlight (Version 0.1.7)
- Seurat (Version 4.0.5)
- STdeconvolve (Version 0.1.0)
- EnDecon
Python
- cell2location (Version 0.7a0, python = 3.7)
- DestVI (scvi-tools Version 0.11.0, python = 3.7)
- stereoscope (Version 0.3.1, python = 3.9.5)
- STRIDE (Version 0.0.1b0, python = 3.8.12)
- Tangram (Version 1.0.0, python = 3.8.13)