[OpenReview] [arXiv] [Code]
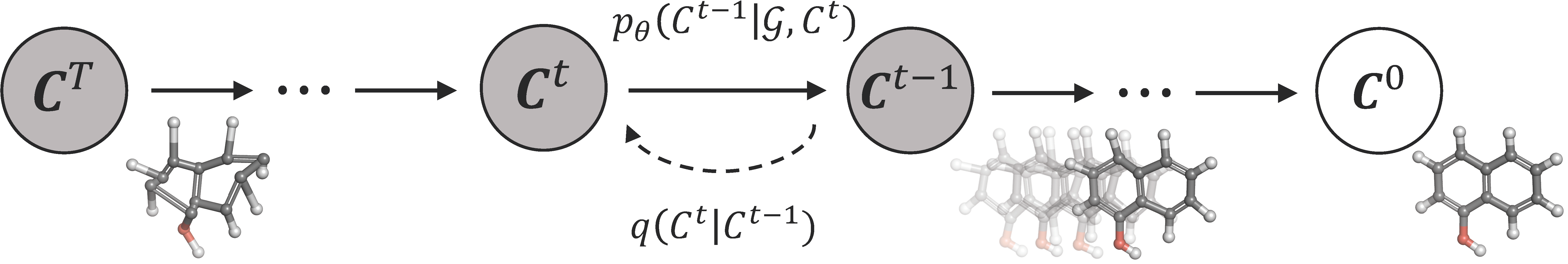
The official implementation of GeoDiff: A Geometric Diffusion Model for Molecular Conformation Generation (ICLR 2022 Oral Presentation [54/3391]).
# Clone the environment
conda env create -f env.yml
# Activate the environment
conda activate geodiff
# Install PyG
conda install pytorch-geometric=1.7.2=py37_torch_1.8.0_cu102 -c rusty1s -c conda-forgeThe offical raw GEOM dataset is avaiable [here].
We provide the preprocessed datasets (GEOM) in this [google drive folder]. After downleading the dataset, it should be put into the folder path as specified in the dataset variable of config files ./configs/*.yml.
You can also download origianl GEOM full dataset and prepare your own data split. A guide is available at previous work ConfGF's [github page].
All hyper-parameters and training details are provided in config files (./configs/*.yml), and free feel to tune these parameters.
You can train the model with the following commands:
# Default settings
python train.py ./configs/qm9_default.yml
python train.py ./configs/drugs_default.yml
# An ablation setting with fewer timesteps, as described in Appendix D.2.
python train.py ./configs/drugs_1k_default.ymlThe model checkpoints, configuration yaml file as well as training log will be saved into a directory specified by --logdir in train.py.
We provide the checkpoints of two trained models, i.e., qm9_default and drugs_default in the [google drive folder]. Note that, please put the checkpoints *.pt into paths like ${log}/${model}/checkpoints/, and also put corresponding configuration file *.yml into the upper level directory ${log}/${model}/.
Attention: if you want to use pretrained models, please use the code at the pretrain branch, which is the vanilla codebase for reproducing the results with our pretrained models. We recently notice some issue of the codebase and update it, making the main branch not compatible well with the previous checkpoints.
You can generate conformations for entire or part of test sets by:
python test.py ${log}/${model}/checkpoints/${iter}.pt \
--start_idx 800 --end_idx 1000Here start_idx and end_idx indicate the range of the test set that we want to use. All hyper-parameters related to sampling can be set in test.py files. Specifically, for testing qm9 model, you could add the additional arg --w_global 0.3, which empirically shows slightly better results.
Conformations of some drug-like molecules generated by GeoDiff are provided below.
After generating conformations following the obove commands, the results of all benchmark tasks can be calculated based on the generated data.
The COV and MAT scores on the GEOM datasets can be calculated using the following commands:
python eval_covmat.py ${log}/${model}/${sample}/sample_all.pklFor the property prediction, we use a small split of qm9 different from the Conformation Generation task. This split is also provided in the [google drive folder]. Generating conformations and evaluate mean absolute errors (MAR) metric on this split can be done by the following commands:
python ${log}/${model}/checkpoints/${iter}.pt --num_confs 50 \
--start_idx 0 --test_set data/GEOM/QM9/qm9_property.pkl
python eval_prop.py --generated ${log}/${model}/${sample}/sample_all.pklHere we also provide a guideline for visualizing molecules with PyMol. The guideline is borrowed from previous work ConfGF's [github page].
pymol -RDisplay - Background - WhiteDisplay - Color Space - CMYKDisplay - Quality - Maximal QualityDisplay Grid- by object: use
set grid_slot, int, mol_nameto put the molecule into the corresponding slot - by state: align all conformations in a single slot
- by object-state: align all conformations and put them in separate slots. (
grid_slotdont work!)
- by object: use
Setting - Line and Sticks - Ball and Stick on - Ball and Stick ratio: 1.5Setting - Line and Sticks - Stick radius: 0.2 - Stick Hydrogen Scale: 1.0
-
To show molecules
hide everythingshow sticks
-
To align molecules:
align name1, name2 -
Convert RDKit mol to Pymol
from rdkit.Chem import PyMol v= PyMol.MolViewer() rdmol = Chem.MolFromSmiles('C') v.ShowMol(rdmol, name='mol') v.SaveFile('mol.pkl')
Please consider citing the our paper if you find it helpful. Thank you!
@inproceedings{
xu2022geodiff,
title={GeoDiff: A Geometric Diffusion Model for Molecular Conformation Generation},
author={Minkai Xu and Lantao Yu and Yang Song and Chence Shi and Stefano Ermon and Jian Tang},
booktitle={International Conference on Learning Representations},
year={2022},
url={https://openreview.net/forum?id=PzcvxEMzvQC}
}
This repo is built upon the previous work ConfGF's [codebase]. Thanks Chence and Shitong!
If you have any question, please contact me at [email protected] or [email protected].
- The current codebase is not compatible with more recent torch-geometric versions.
- The current processed dataset (with PyD data object) is not compatible with more recent torch-geometric versions.