rISIMIP is an R package for accessing and
analysing data provided by the Inter-sectoral Impact Model
Intercomparison Project (ISIMIP). Data from the different simulation
rounds (ISIMIP2a, ISIMIP2b, ISIMIP3a, ISIMIP3b) is available from
here. For more information
on the different data types and data input and output products have a
look at the ISIMIP Website. The package
currently consists of two functions:
readISIMIP()reads and pre-processes ISIMIP datalistISIMIP()creates a list of requested ISIMIP data files
You can learn more about them in vignette("rISIMIP").
An example of extracting country-specific data from ISIMIP2b can be
found in vignette("country-specific").
To use the package, install it directly from GitHub using the
remotes package:
# Install remotes if not previously installed
if(!"remotes" %in% installed.packages()[,"Package"]) install.packages("remotes")
# Install rISIMIP from Github if not previously installed
if(!"rISIMIP" %in% installed.packages()[,"Package"]) remotes::install_github("RS-eco/rISIMIP", build_vignettes = TRUE)If you encounter a bug or if you have any problems, please file an issue on Github.
# Load rISIMIP package
library(rISIMIP)The function listISIMIP just lists all climate files for the desired
time period, model and variable. The files can then be put into the
aggregateNC function of the processNC package for processing the
required NetCDF files.
# List urban area files for histsoc scenario - ISIMIP2b
listISIMIP(path="I:/", version="ISIMIP2b", type="landuse",
scenario="histsoc", var="urbanareas", startyear=1861, endyear=2005)
# List crop data files for histsoc scenario - ISIMIP3b
listISIMIP(path="I:/", version="ISIMIP3b", type="landuse",
scenario="histsoc", var="5crops", startyear=1861, endyear=2005)Note: The path must lead to a file directory on your computer, which contains the required ISIMIP files. You can download the required ISIMIP data files from: https://esg.pik-potsdam.de/search/isimip/
With readISIMIP you can read one or multiple ISIMIP datafiles into a
raster stack.
# Read urban area file for 2005soc scenario - ISIMIP2b
(urbanareas_1970_1999 <- readISIMIP(path="I:/", type="landuse", scenario="2005soc",
var="urbanareas", startyear=1970, endyear=1999))
# Read pasture file for 2015soc scenario - ISIMIP3b
(pastures_1970_1999 <- readISIMIP(path="I:/", type="landuse", scenario="2015soc",
var="pastures", startyear=1970, endyear=1999))However, this is not useful if you are interested in long time periods, as one datafile is about 7 GB in size and you will quickly run into memory limitations.
rISIMIP contains various pre-processed data. See the data-raw folder
for how we derived the included datasets.
Annual global mean temperature as well as the 31-year running mean were
calculated for each GCM and four RCPs (RCP2.6, RCP4.5, RCP6.0 and
RCP8.5). Furthermore, the year when the 31-year runnning mean of global
mean temperature crosses a certain temperature threshold has been
calculated. The data has been provided by ISIMIP and a summary of it can
be accessed from the vignette("temperature-thresholds") vignette and
is also available from the ISIMIP
Website.
The landseamask used by ISIMIP has been included in the package and can be accessed by:
data("landseamask_generic")The code for calculating global bioclimatic data from ISIMIP2b and
ISIMIP3b mnodel output can be found in vignette("global-landonly") and
vignette("global-landonly-isimip3b") respectively.
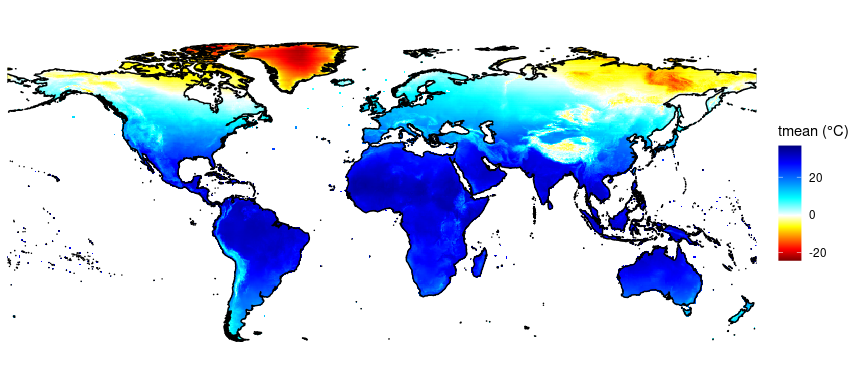
Current and future bioclimatic data for three 30-yr periods (1995, 2050, 2080) was derived from the EWEMBI (https://esg.pik-potsdam.de/search/isimip/?project=ISIMIP2b&product=input_secondary&dataset_type=Climate+atmosphere+observed) and ISIMIP2b data (https://esg.pik-potsdam.de/search/isimip/?project=ISIMIP2b&product=input&dataset_type=Climate+atmosphere+simulated) and is included in this package.
EWEMBI - 1995
data("bioclim_ewembi_1995_landonly")
library(dplyr); library(sf); library(ggplot2)
data(outline, package="ggmap2")
outline <- sf::st_as_sf(outline)
col_val <- scales::rescale(unique(c(seq(min(bioclim_ewembi_1995_landonly$bio1), 0, length=5),
seq(0, max(bioclim_ewembi_1995_landonly$bio1), length=5))))
bioclim_ewembi_1995_landonly %>% select(x,y,bio1) %>%
ggplot() + geom_tile(aes(x=x, y=y, fill=bio1)) +
geom_sf(data=outline, fill="transparent", colour="black") +
scale_fill_gradientn(name="tmean (°C)", colours=rev(colorRampPalette(
c("#00007F", "blue", "#007FFF", "cyan",
"white", "yellow", "#FF7F00", "red", "#7F0000"))(255)),
na.value="transparent", values=col_val,
limits=c(min(bioclim_ewembi_1995_landonly$bio1)-2,
max(bioclim_ewembi_1995_landonly$bio1)+2)) +
coord_sf(expand=F,
xlim=c(min(bioclim_ewembi_1995_landonly$x),
max(bioclim_ewembi_1995_landonly$x)),
ylim=c(min(bioclim_ewembi_1995_landonly$y),
max(bioclim_ewembi_1995_landonly$y)),
ndiscr=0) + theme_classic() +
theme(axis.title = element_blank(), axis.line = element_blank(),
axis.ticks = element_blank(), axis.text = element_blank(),
plot.background = element_rect(fill = "transparent"),
legend.background = element_rect(fill = "transparent"),
legend.box.background = element_rect(fill = "transparent", colour=NA))RCP2.6 - 2080
data("bioclim_gfdl-esm2m_rcp26_2080_landonly")
data("bioclim_hadgem2-es_rcp26_2080_landonly")
data("bioclim_ipsl-cm5a-lr_rcp26_2080_landonly")
data("bioclim_miroc5_rcp26_2080_landonly")
bioclim_rcp26_2080_landonly <- bind_rows(`bioclim_gfdl-esm2m_rcp26_2080_landonly`,
`bioclim_hadgem2-es_rcp26_2080_landonly`,
`bioclim_ipsl-cm5a-lr_rcp26_2080_landonly`,
`bioclim_miroc5_rcp26_2080_landonly`) %>%
select(x,y,bio1) %>% group_by(x,y) %>% summarise(bio1=mean(bio1, na.rm=T))
col_val <- scales::rescale(unique(c(seq(min(bioclim_rcp26_2080_landonly$bio1), 0, length=5),
seq(0, max(bioclim_rcp26_2080_landonly$bio1), length=5))))
ggplot() + geom_tile(data=bioclim_rcp26_2080_landonly, aes(x=x, y=y, fill=bio1)) +
geom_sf(data=outline, fill="transparent", colour="black") +
scale_fill_gradientn(name="tmean (°C)", colours=rev(colorRampPalette(
c("#00007F", "blue", "#007FFF", "cyan",
"white", "yellow", "#FF7F00", "red", "#7F0000"))(255)),
na.value="transparent", values=col_val,
limits=c(min(bioclim_rcp26_2080_landonly$bio1)-2,
max(bioclim_rcp26_2080_landonly$bio1)+2)) +
coord_sf(expand=F,
xlim=c(min(bioclim_rcp26_2080_landonly$x),
max(bioclim_rcp26_2080_landonly$x)),
ylim=c(min(bioclim_rcp26_2080_landonly$y),
max(bioclim_rcp26_2080_landonly$y)),
ndiscr=0) + theme_classic() +
theme(axis.title = element_blank(), axis.line = element_blank(),
axis.ticks = element_blank(), axis.text = element_blank(),
plot.background = element_rect(fill = "transparent"),
legend.background = element_rect(fill = "transparent"),
legend.box.background = element_rect(fill = "transparent", colour=NA))RCP6.0 - 2080
data("bioclim_gfdl-esm2m_rcp60_2080_landonly")
data("bioclim_hadgem2-es_rcp60_2080_landonly")
data("bioclim_ipsl-cm5a-lr_rcp60_2080_landonly")
data("bioclim_miroc5_rcp60_2080_landonly")
bioclim_rcp60_2080_landonly <- bind_rows(`bioclim_gfdl-esm2m_rcp60_2080_landonly`,
`bioclim_hadgem2-es_rcp60_2080_landonly`,
`bioclim_ipsl-cm5a-lr_rcp60_2080_landonly`,
`bioclim_miroc5_rcp60_2080_landonly`) %>%
select(x,y,bio1) %>% group_by(x,y) %>% summarise(bio1=mean(bio1, na.rm=T))
col_val <- scales::rescale(unique(c(seq(min(bioclim_rcp60_2080_landonly$bio1), 0, length=5),
seq(0, max(bioclim_rcp60_2080_landonly$bio1), length=5))))
ggplot() + geom_tile(data=bioclim_rcp60_2080_landonly, aes(x=x, y=y, fill=bio1)) +
geom_sf(data=outline, fill="transparent", colour="black") +
scale_fill_gradientn(name="tmean (°C)", colours=rev(colorRampPalette(
c("#00007F", "blue", "#007FFF", "cyan",
"white", "yellow", "#FF7F00", "red", "#7F0000"))(255)),
na.value="transparent", values=col_val,
limits=c(min(bioclim_rcp60_2080_landonly$bio1)-2,
max(bioclim_rcp60_2080_landonly$bio1)+2)) +
coord_sf(expand=F,
xlim=c(min(bioclim_rcp60_2080_landonly$x),
max(bioclim_rcp60_2080_landonly$x)),
ylim=c(min(bioclim_rcp60_2080_landonly$y),
max(bioclim_rcp60_2080_landonly$y)),
ndiscr=0) + theme_classic() +
theme(axis.title = element_blank(), axis.line = element_blank(),
axis.ticks = element_blank(), axis.text = element_blank(),
plot.background = element_rect(fill = "transparent"),
legend.background = element_rect(fill = "transparent"),
legend.box.background = element_rect(fill = "transparent", colour=NA))RCP8.5 - 2080
data("bioclim_gfdl-esm2m_rcp85_2080_landonly")
data("bioclim_hadgem2-es_rcp85_2080_landonly")
data("bioclim_ipsl-cm5a-lr_rcp85_2080_landonly")
data("bioclim_miroc5_rcp85_2080_landonly")
bioclim_rcp85_2080_landonly <- bind_rows(`bioclim_gfdl-esm2m_rcp85_2080_landonly`,
`bioclim_hadgem2-es_rcp85_2080_landonly`,
`bioclim_ipsl-cm5a-lr_rcp85_2080_landonly`,
`bioclim_miroc5_rcp85_2080_landonly`) %>%
select(x,y,bio1) %>% group_by(x,y) %>% summarise(bio1=mean(bio1, na.rm=T))
col_val <- scales::rescale(unique(c(seq(min(bioclim_rcp85_2080_landonly$bio1), 0, length=5),
seq(0, max(bioclim_rcp85_2080_landonly$bio1), length=5))))
ggplot() + geom_tile(data=bioclim_rcp85_2080_landonly, aes(x=x, y=y, fill=bio1)) +
geom_sf(data=outline, fill="transparent", colour="black") +
scale_fill_gradientn(name="tmean (°C)", colours=rev(colorRampPalette(
c("#00007F", "blue", "#007FFF", "cyan",
"white", "yellow", "#FF7F00", "red", "#7F0000"))(255)),
na.value="transparent", values=col_val,
limits=c(min(bioclim_rcp85_2080_landonly$bio1)-2,
max(bioclim_rcp85_2080_landonly$bio1)+2)) +
coord_sf(expand=F,
xlim=c(min(bioclim_rcp85_2080_landonly$x),
max(bioclim_rcp85_2080_landonly$x)),
ylim=c(min(bioclim_rcp85_2080_landonly$y),
max(bioclim_rcp85_2080_landonly$y)),
ndiscr=0) + theme_classic() +
theme(axis.title = element_blank(), axis.line = element_blank(),
axis.ticks = element_blank(), axis.text = element_blank(),
plot.background = element_rect(fill = "transparent"),
legend.background = element_rect(fill = "transparent"),
legend.box.background = element_rect(fill = "transparent", colour=NA))Current and future bioclimatic data for five 30-yr periods (1995, 2000, 2005, 2050, 2080) was derived from the GSWP3-W5E5 and ISIMIP3b data (https://esg.pik-potsdam.de/search/isimip/?project=ISIMIP3b&product=input&dataset_type=Climate+atmosphere+simulated) and is included in this package.
GSWP3_W5E5 - 2005
data("bioclim_gswp3-w5e5_obsclim_2005_landonly")
library(dplyr); library(sf); library(ggplot2)
data(outline, package="ggmap2")
outline <- sf::st_as_sf(outline)
col_val <- scales::rescale(unique(c(seq(min(`bioclim_gswp3-w5e5_obsclim_2005_landonly`$bio1), 0, length=5),
seq(0, max(`bioclim_gswp3-w5e5_obsclim_2005_landonly`$bio1), length=5))))
`bioclim_gswp3-w5e5_obsclim_2005_landonly` %>% select(x,y,bio1) %>%
ggplot() + geom_tile(aes(x=x, y=y, fill=bio1)) +
geom_sf(data=outline, fill="transparent", colour="black") +
scale_fill_gradientn(name="tmean (°C)", colours=rev(colorRampPalette(
c("#00007F", "blue", "#007FFF", "cyan",
"white", "yellow", "#FF7F00", "red", "#7F0000"))(255)),
na.value="transparent", values=col_val,
limits=c(min(`bioclim_gswp3-w5e5_obsclim_2005_landonly`$bio1)-2,
max(`bioclim_gswp3-w5e5_obsclim_2005_landonly`$bio1)+2)) +
coord_sf(expand=F,
xlim=c(min(`bioclim_gswp3-w5e5_obsclim_2005_landonly`$x),
max(`bioclim_gswp3-w5e5_obsclim_2005_landonly`$x)),
ylim=c(min(`bioclim_gswp3-w5e5_obsclim_2005_landonly`$y),
max(`bioclim_gswp3-w5e5_obsclim_2005_landonly`$y)),
ndiscr=0) + theme_classic() +
theme(axis.title = element_blank(), axis.line = element_blank(),
axis.ticks = element_blank(), axis.text = element_blank(),
plot.background = element_rect(fill = "transparent"),
legend.background = element_rect(fill = "transparent"),
legend.box.background = element_rect(fill = "transparent", colour=NA))SSP126 - 2080
data("bioclim_gfdl-esm4_ssp126_2080_landonly")
data("bioclim_ipsl-cm6a-lr_ssp126_2080_landonly")
data("bioclim_mpi-esm1-2-hr_ssp126_2080_landonly")
data("bioclim_mri-esm2-0_ssp126_2080_landonly")
data("bioclim_ukesm1-0-ll_ssp126_2080_landonly")
bioclim_ssp126_2080_landonly <- bind_rows(`bioclim_gfdl-esm4_ssp126_2080_landonly`,
`bioclim_ipsl-cm6a-lr_ssp126_2080_landonly`,
`bioclim_mpi-esm1-2-hr_ssp126_2080_landonly`,
`bioclim_mri-esm2-0_ssp126_2080_landonly`,
`bioclim_ukesm1-0-ll_ssp126_2080_landonly`) %>%
select(x,y,bio1) %>% group_by(x,y) %>% summarise(bio1=mean(bio1, na.rm=T))
col_val <- scales::rescale(unique(c(seq(min(bioclim_ssp126_2080_landonly$bio1), 0, length=5),
seq(0, max(bioclim_ssp126_2080_landonly$bio1), length=5))))
ggplot() + geom_tile(data=bioclim_ssp126_2080_landonly, aes(x=x, y=y, fill=bio1)) +
geom_sf(data=outline, fill="transparent", colour="black") +
scale_fill_gradientn(name="tmean (°C)", colours=rev(colorRampPalette(
c("#00007F", "blue", "#007FFF", "cyan",
"white", "yellow", "#FF7F00", "red", "#7F0000"))(255)),
na.value="transparent", values=col_val,
limits=c(min(bioclim_ssp126_2080_landonly$bio1)-2,
max(bioclim_ssp126_2080_landonly$bio1)+2)) +
coord_sf(expand=F,
xlim=c(min(bioclim_ssp126_2080_landonly$x),
max(bioclim_ssp126_2080_landonly$x)),
ylim=c(min(bioclim_ssp126_2080_landonly$y),
max(bioclim_ssp126_2080_landonly$y)),
ndiscr=0) + theme_classic() +
theme(axis.title = element_blank(), axis.line = element_blank(),
axis.ticks = element_blank(), axis.text = element_blank(),
plot.background = element_rect(fill = "transparent"),
legend.background = element_rect(fill = "transparent"),
legend.box.background = element_rect(fill = "transparent", colour=NA))SSP370 - 2080
data("bioclim_gfdl-esm4_ssp370_2080_landonly")
data("bioclim_ipsl-cm6a-lr_ssp370_2080_landonly")
data("bioclim_mpi-esm1-2-hr_ssp370_2080_landonly")
data("bioclim_mri-esm2-0_ssp370_2080_landonly")
data("bioclim_ukesm1-0-ll_ssp370_2080_landonly")
bioclim_ssp370_2080_landonly <- bind_rows(`bioclim_gfdl-esm4_ssp370_2080_landonly`,
`bioclim_ipsl-cm6a-lr_ssp370_2080_landonly`,
`bioclim_mpi-esm1-2-hr_ssp370_2080_landonly`,
`bioclim_mri-esm2-0_ssp370_2080_landonly`,
`bioclim_ukesm1-0-ll_ssp370_2080_landonly`) %>%
select(x,y,bio1) %>% group_by(x,y) %>% summarise(bio1=mean(bio1, na.rm=T))
col_val <- scales::rescale(unique(c(seq(min(bioclim_ssp370_2080_landonly$bio1), 0, length=5),
seq(0, max(bioclim_ssp370_2080_landonly$bio1), length=5))))
ggplot() + geom_tile(data=bioclim_ssp370_2080_landonly, aes(x=x, y=y, fill=bio1)) +
geom_sf(data=outline, fill="transparent", colour="black") +
scale_fill_gradientn(name="tmean (°C)", colours=rev(colorRampPalette(
c("#00007F", "blue", "#007FFF", "cyan",
"white", "yellow", "#FF7F00", "red", "#7F0000"))(255)),
na.value="transparent", values=col_val,
limits=c(min(bioclim_ssp370_2080_landonly$bio1)-2,
max(bioclim_ssp370_2080_landonly$bio1)+2)) +
coord_sf(expand=F,
xlim=c(min(bioclim_ssp370_2080_landonly$x),
max(bioclim_ssp370_2080_landonly$x)),
ylim=c(min(bioclim_ssp370_2080_landonly$y),
max(bioclim_ssp370_2080_landonly$y)),
ndiscr=0) + theme_classic() +
theme(axis.title = element_blank(), axis.line = element_blank(),
axis.ticks = element_blank(), axis.text = element_blank(),
plot.background = element_rect(fill = "transparent"),
legend.background = element_rect(fill = "transparent"),
legend.box.background = element_rect(fill = "transparent", colour=NA))SSP585 - 2080
data("bioclim_gfdl-esm4_ssp585_2080_landonly")
data("bioclim_ipsl-cm6a-lr_ssp585_2080_landonly")
data("bioclim_mpi-esm1-2-hr_ssp585_2080_landonly")
data("bioclim_mri-esm2-0_ssp585_2080_landonly")
data("bioclim_ukesm1-0-ll_ssp585_2080_landonly")
bioclim_ssp585_2080_landonly <- bind_rows(`bioclim_gfdl-esm4_ssp585_2080_landonly`,
`bioclim_ipsl-cm6a-lr_ssp585_2080_landonly`,
`bioclim_mpi-esm1-2-hr_ssp585_2080_landonly`,
`bioclim_mri-esm2-0_ssp585_2080_landonly`,
`bioclim_ukesm1-0-ll_ssp585_2080_landonly`) %>%
select(x,y,bio1) %>% group_by(x,y) %>% summarise(bio1=mean(bio1, na.rm=T))
col_val <- scales::rescale(unique(c(seq(min(bioclim_ssp585_2080_landonly$bio1), 0, length=5),
seq(0, max(bioclim_ssp585_2080_landonly$bio1), length=5))))
ggplot() + geom_tile(data=bioclim_ssp585_2080_landonly, aes(x=x, y=y, fill=bio1)) +
geom_sf(data=outline, fill="transparent", colour="black") +
scale_fill_gradientn(name="tmean (°C)", colours=rev(colorRampPalette(
c("#00007F", "blue", "#007FFF", "cyan",
"white", "yellow", "#FF7F00", "red", "#7F0000"))(255)),
na.value="transparent", values=col_val,
limits=c(min(bioclim_ssp585_2080_landonly$bio1)-2,
max(bioclim_ssp585_2080_landonly$bio1)+2)) +
coord_sf(expand=F,
xlim=c(min(bioclim_ssp585_2080_landonly$x),
max(bioclim_ssp585_2080_landonly$x)),
ylim=c(min(bioclim_ssp585_2080_landonly$y),
max(bioclim_ssp585_2080_landonly$y)),
ndiscr=0) + theme_classic() +
theme(axis.title = element_blank(), axis.line = element_blank(),
axis.ticks = element_blank(), axis.text = element_blank(),
plot.background = element_rect(fill = "transparent"),
legend.background = element_rect(fill = "transparent"),
legend.box.background = element_rect(fill = "transparent", colour=NA))