Implementation of the paper:
GPC: Generative and general pathology image classifier
Anh Tien Nguyen and Jin Tae Kwak
MedAGI: MICCAI 2023 1st International Workshop on Foundation Models for General Medical AI
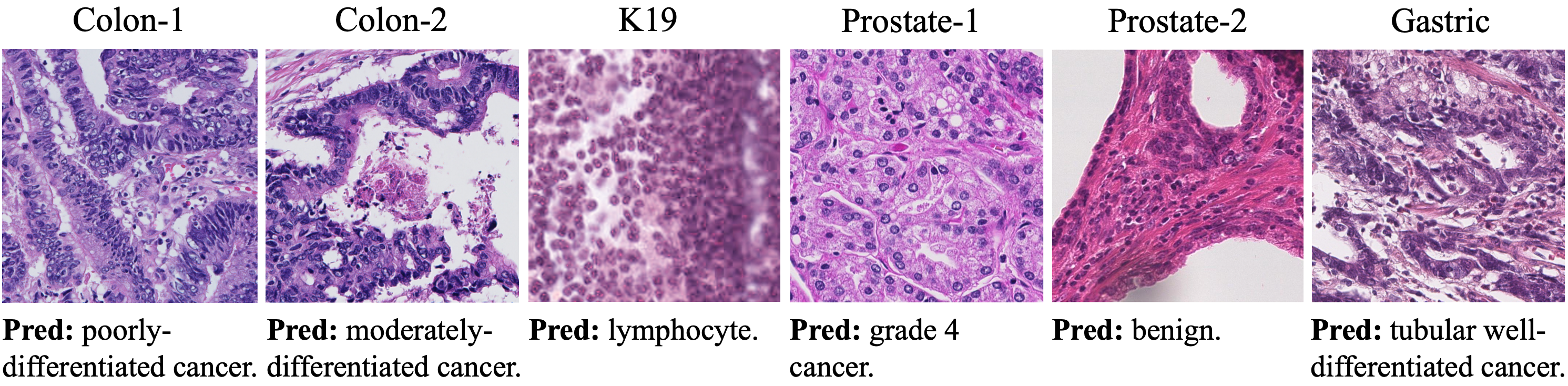
Deep learning has been increasingly incorporated into many computational pathology applications to improve its efficiency, accuracy, and robustness. Although successful, most previous approaches of using convolutional neural networks and Transformers have crucial drawbacks. There exist numerous tasks in pathology, but one needs to build a model per task, i.e., a task-specific model, thereby increasing the number of models, training resources, and cost. Moreover, transferring any task- specific model to another task is still a challenging problem. Herein, we propose a task-agnostic generative and general pathology image classifier, so called GPC, that aims at processing and learning from any kinds of pathology images as well as conducting any pathology classification tasks. Equipped with a convolutional neural network and a Transformer-based language model, GPC maps pathology images into a high-dimensional feature space and generates pertinent class labels as texts via the image- to-text classification mechanism. We evaluate GPC on six datasets for four different pathology image classification tasks. Experimental results show that GPC holds considerable potential for developing an effective and efficient universal model for pathology image analysis.
git clone https://github.com/QuIIL/gpc
cd gpc
conda create --name gpc --file requirements.txt
conda activate gpc
pip install -r requirements.txt
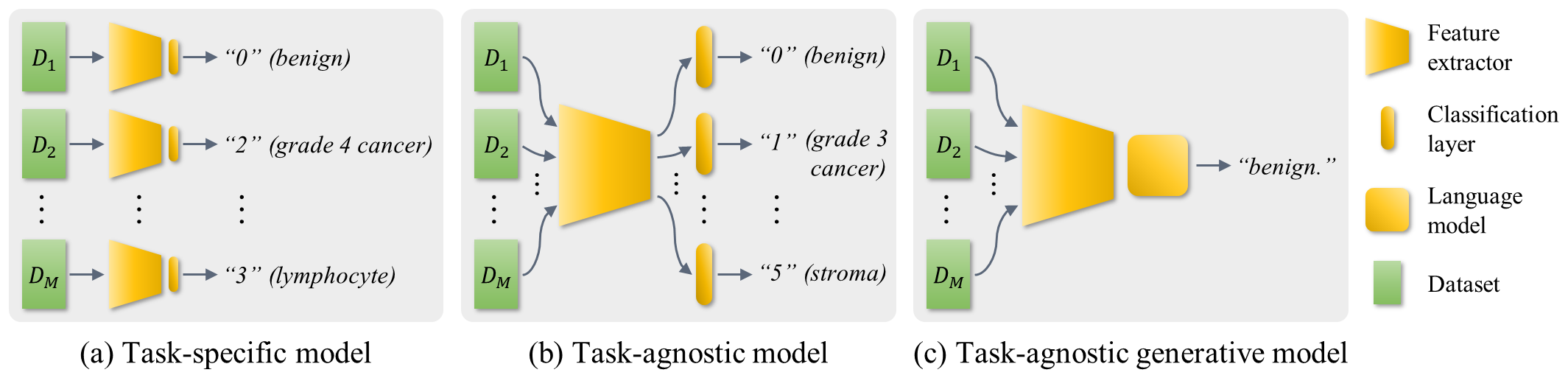
- Task-specific: models are trained on a training set and tested on the test set(s) per classification task
- Task-agnostic: models trained on all training sets from four classification tasks and assessed on each test set per task using the corresponding classifier (use only the output of the corresponding branch of testing task)
- Task-agnostic generative: model trained on all training sets from four classification tasks and evaluated on the entire test sets
Training GPC with DDP:
CUDA_VISIBLE_DEVICES=0,1 python train.py \
--encoder convnext_large
--lm facebook/opt-125m
--out_dir <your_dir>
Training a task-specific CNN:
python experiment/train_cnn.py \
--cnn_type resnet50
--dataset uhu
--out_dir <your_dir>
Training a task-agnostic CNN (multi-branch):
python experiment/train_multi_branch.py \
--cnn_type resnet50
--dataset prostate_1
--out_dir <your_dir>
Testing GPC with DDP:
CUDA_VISIBLE_DEVICES=0,1 python test.py \
--dataset prostate_2
--pretrain_path <ckpt_path>
Testing a task-specific CNN:
python experiment/test_cnn.py \
--dataset prostate_2
--pretrain_path <ckpt_path>
Testing a task-agnostic CNN (multi-branch):
python experiment/test_multi_branch.py \
--dataset prostate_2
--pretrain_path <ckpt_path>