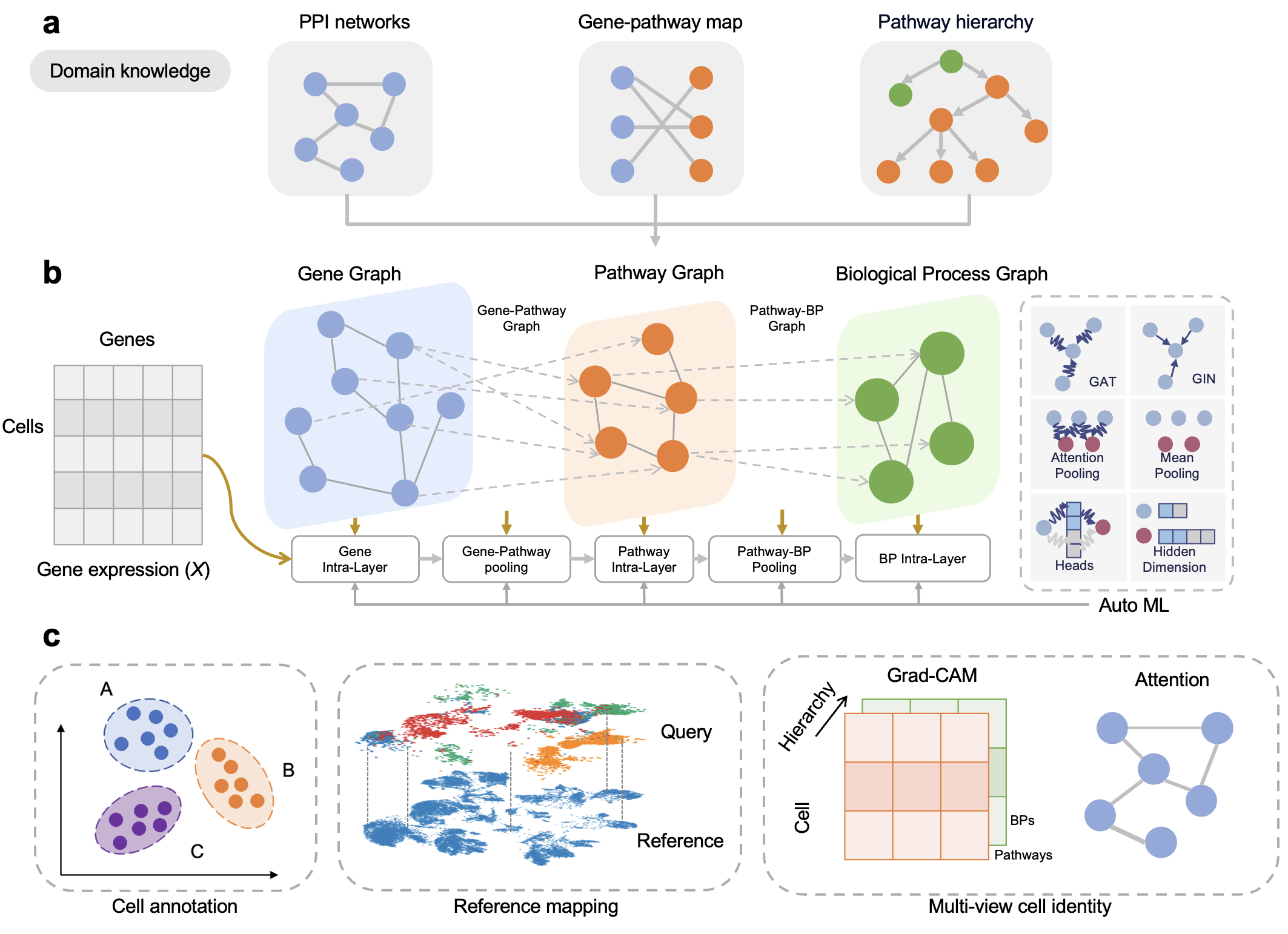
We created the python package called celldecoder that that decoding cell identity from gene expressions by explicitly modeling the multi-scale biological interactions, i.e., genes, pathways, and biological processes.
- Python >= 3.8
- torch == 2.0.1
- torch-geometric == 2.3.1
- CUDA 11.7
conda create -n celldecoder python=3.8
conda activate celldecoder
conda install pytorch=2.0.1 cudatoolkit=11.7 -c pytorch
pip install torch_geometric
pip install torch_scatter torch_sparse torch_cluster -f https://data.pyg.org/whl/torch-2.0.0+cu117.html
The celldecoder python package is in the folder celldecoder. You can simply install it from the root of this repository using
pip install .
environment.yml
Building the model requires the use of prior knowledge. Ensure the creation of 2 directories at the software's location: /data/ppi and /data/reactome. The corresponding data can be downloaded from https://figshare.com/articles/dataset/PPI_data/24921831.
hierarchy: gene-pathway mapping & hierarchy pathway informationppi: protein-protein interactions network
from celldecoder.data import interactions,hierarchy
import json
#hierarchy
n_layers = 3 # n layers of the model
reactome = hierarchy.hierarchy_layer(species='HSA') #HSA: human,MMU:mouse
layers = reactome.get_layers(n_levels=n_layers)
ref_adata.uns['hierarchy'] = json.dumps(layers)
query_adata.uns['hierarchy'] = json.dumps(layers)
#ppi
ref_adata = interactions.data_mapping_ppi(ref_adata,ppi_data) #ppi_data: ppi network-
data: anAnnDataobject of reference data and query data (checkout reference and query have the same feature) -
ppi_data: pre-prepared ppi networks data -
./data/ppi: human & mouse processed ppi data -
./data/reactome: hierarchy pathway information
import celldecoder
dataset = "./data/hBone/hBone_ref_adata.h5ad"
device_id = 1
log_dir = f"./log/{dataset}"
# Train the cell decoder using the specified dataset `dataset` on the device `device_id`,
# logging to the specified directory `log_dir`, and using the `cell_label` parameter to specify the cell type label.
celldecoder.train(dataset=dataset, device_id=device_id, log_dir=log_dir, cell_label="cell_type")dataset: dataset namelog_dir: logging directorydevice_id: gpu device id
See other arguments in celldecoder/config.py
./log_dir/args.json: configuration file./log_dir/best.pth: best checkpoint weights
See command output for validation metrics.
import celldecoder
device_id = 1
log_dir = f"./log/hBone"
dataset = "./data/hBone/hBone_query_adata.h5ad"
fn_process = "processed-test"
predict_type = 'cell'
cells = celldecoder.predict(dataset = dataset, device_id = device_id ,log_dir = log_dir, fn_process = fn_process, predict_type = predict_type)dataset: dataset namelog_dir: logging directorydevice_id: gpu device id
See command output for test metrics.
import celldecoder
device_id = 1
log_dir = f"./log/hBone"
dataset = "./data/hBone/hBone_query_adata.h5ad"
fn_process = "processed-test"
celldecoder.embed(dataset = dataset, device_id = device_id ,log_dir = log_dir, out_embed = "output", fn_process = fn_process)dataset: dataset namelog_dir: logging directorydevice_id: gpu device id
Embeddings of the cells
import celldecoder
device_id = 1
log_dir = f"./log/hBone"
dataset = "./data/hBone/hBone_query_adata.h5ad"
fn_process = "processed-test"
celldecoder.explain_feature(dataset = dataset, device_id = device_id ,log_dir = log_dir, explain_method = "grad", fn_process = fn_process)dataset: dataset namelog_dir: logging directorydevice_id: gpu device id
Feature explanations of the cells
import celldecoder
device_id = 1
log_dir = f"./log/hBone"
dataset = "./data/hBone/hBone_query_adata.h5ad"
fn_process = "processed-test"
exp_dict ={
"correlation": 0,
"multi_atten": 1,
"train_sample_gt": 0,
"ce_loss_gt": 0,
"exp_train_epochs": 100,
"exp_lr": 0.01,
}
celldecoder.explain_ppi(dataset = dataset, device_id = device_id ,log_dir = log_dir, fn_process = fn_process, **exp_dict)dataset: dataset namelog_dir: logging directorydevice_id: gpu device id
PPI explanations of the cells
Zhu et al. Decoding cell identity with multi-scale explainable deep learning. bioRxiv 2024.02.05.578922