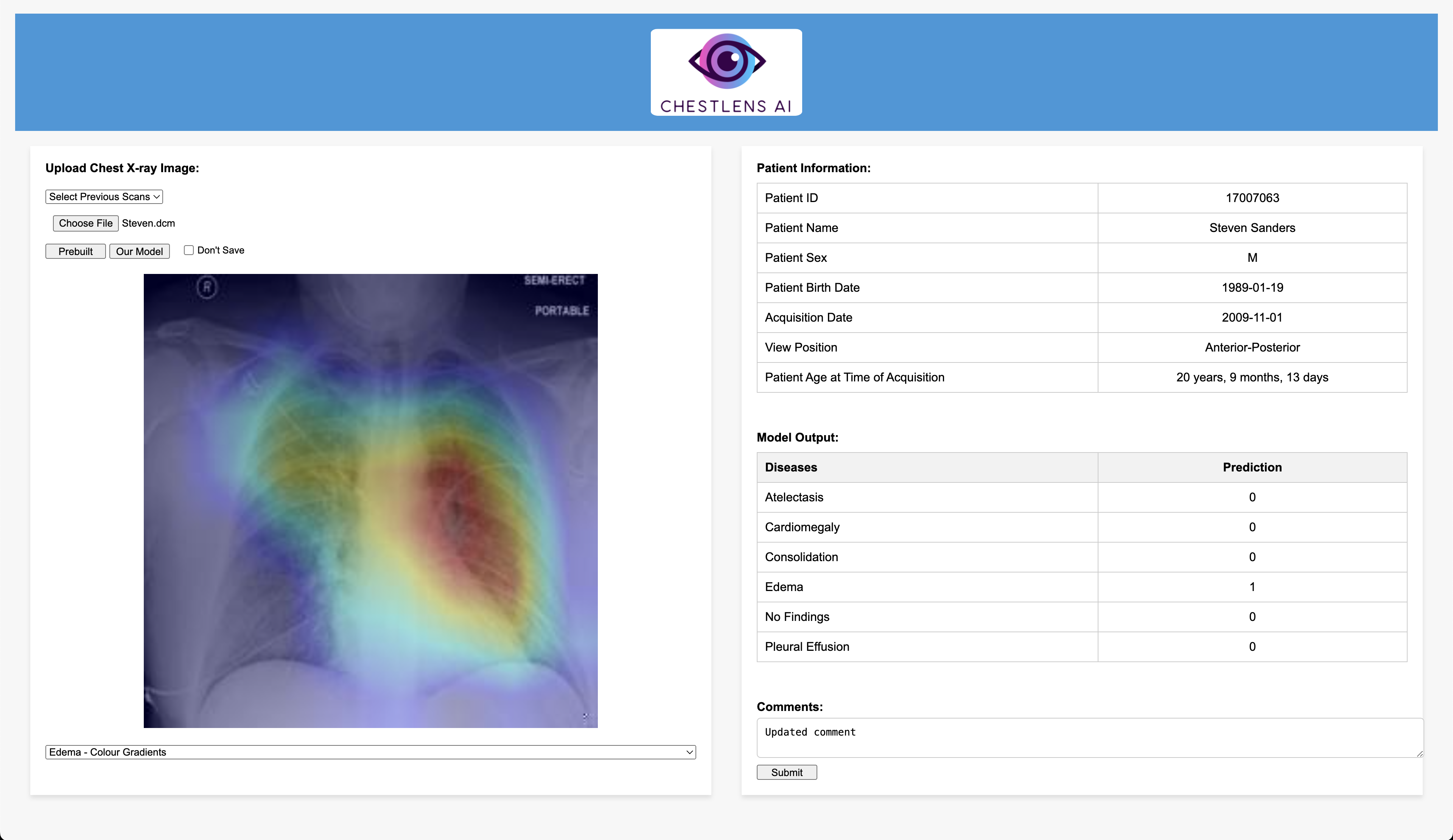
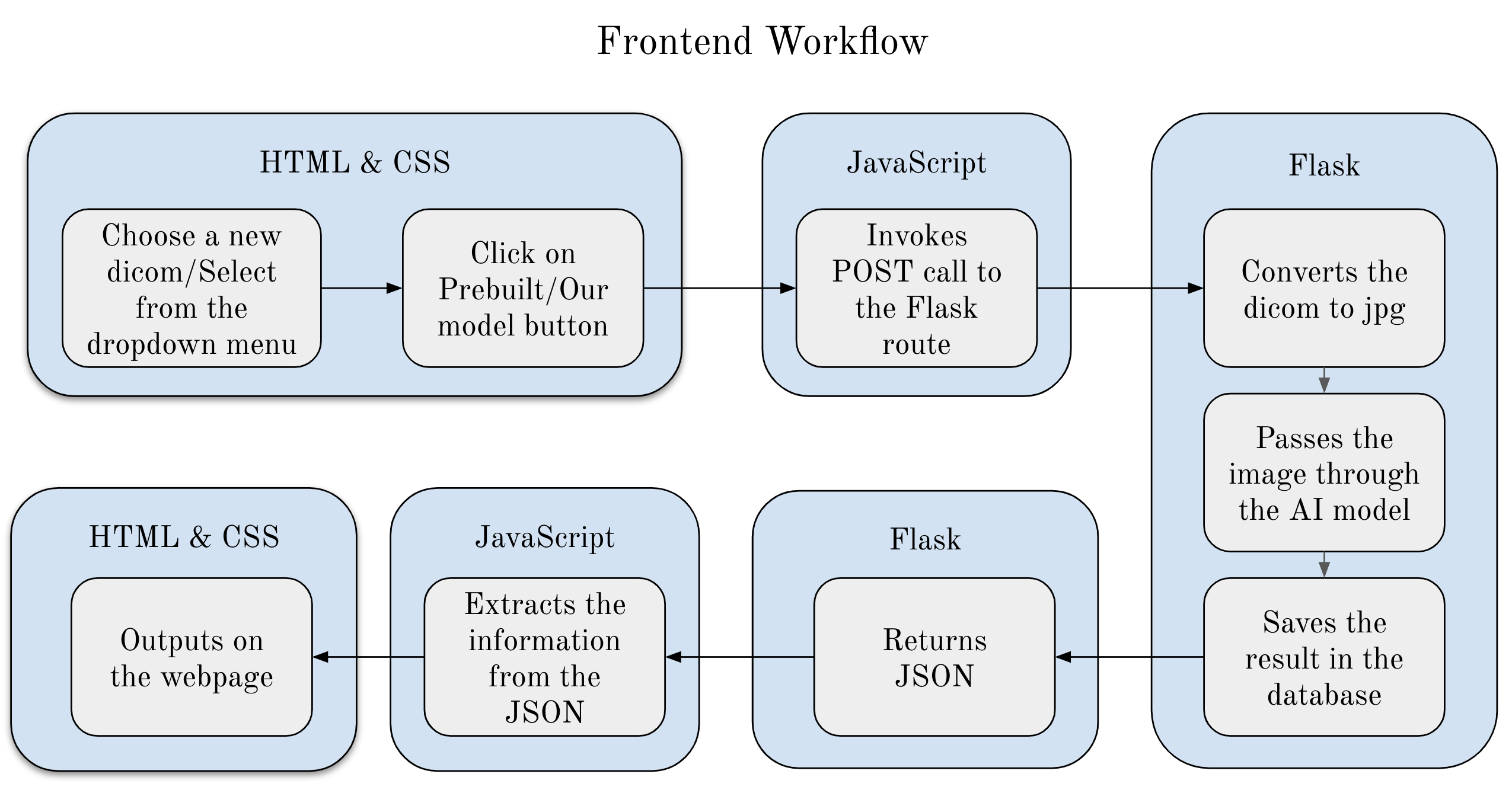
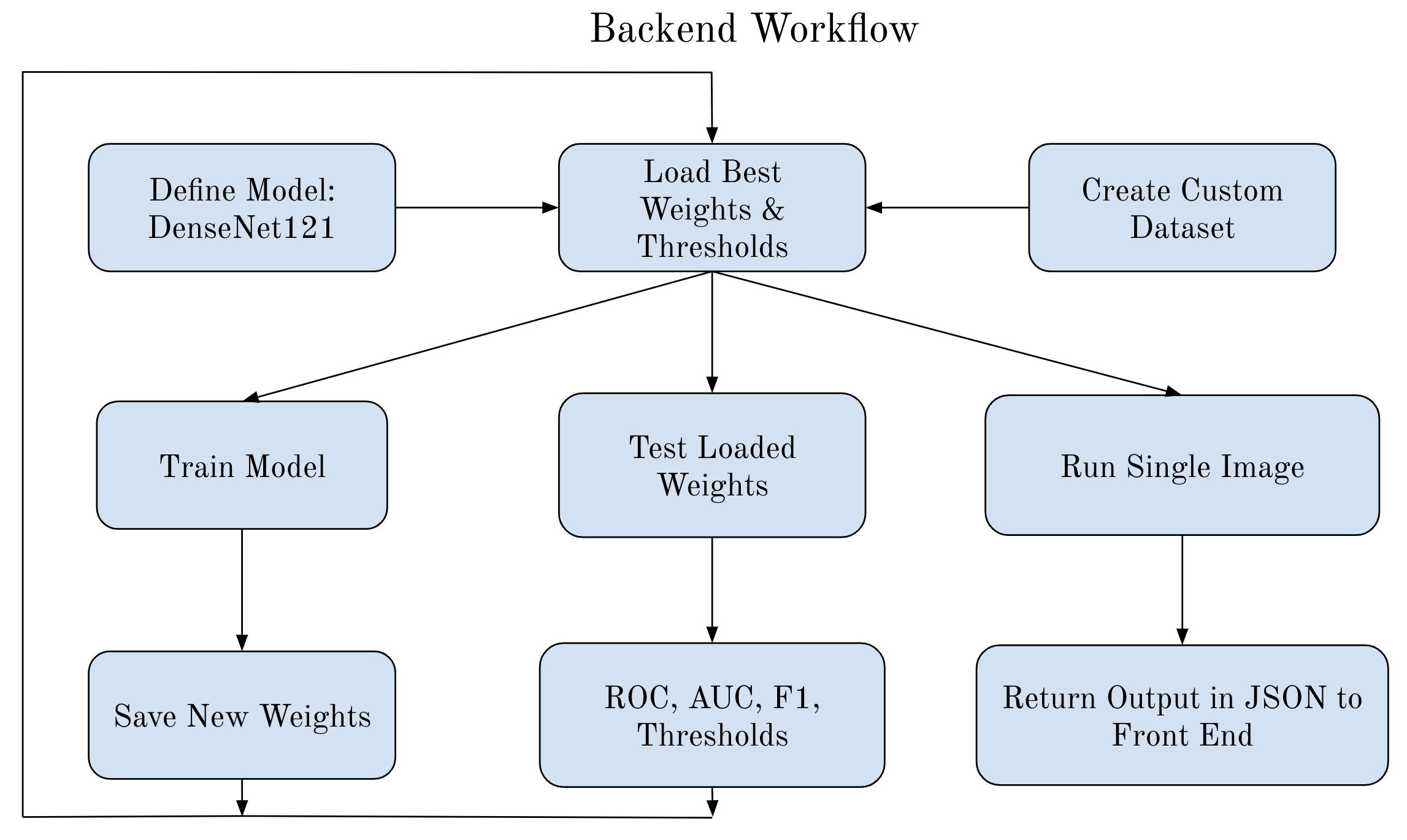
Currently, radiologists manually analyze chest X-ray images to find any abnormalities, injuries, or diseases. This can be very time-consuming and like all things has the possibility of errors. To mitigate this issue we implemented an AI model to identify six common findings. These include Atelectasis, Cardiomegaly, Consolidation, Edema, No Finding, and Pleural Effusion. The main objective of using an AI model on chest X-rays is to help radiologists identify negative results or identify which of these diseases are present and the location of these diseases with the use of visual mapping. The model we used is Densenet121 and was trained in PyTorch using Adam optimizer with a batch size of 64 and the loss function BCE with Logits Loss.
-
Clone this repository.
-
Install the required dependencies.
bash < packages.txt is recommended to maintain Python library versions
bash < packages.txtor
install_packages.bat
or
pip install -r requirements.txt
-
Navigate to the client folder.
cd src/client -
Run the program.
python3 index.py
-
Demo username: [email protected]
-
Demo password (not secure): password3
-
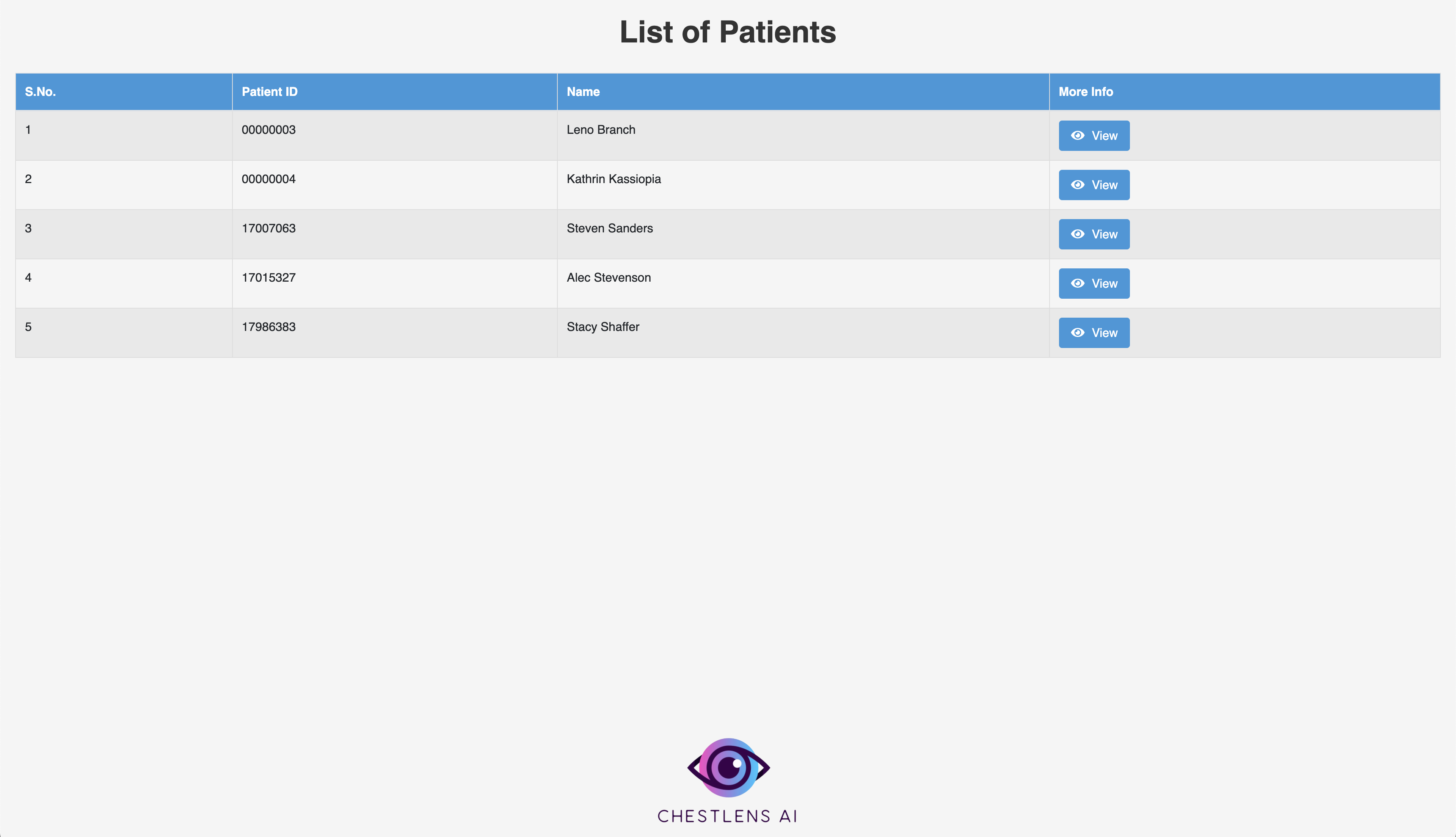
Note: The demo will show six patients, each of which has a corresponding DICOM image available in src/demo_images. Example: src/demo_images/Steven.dcm. Currently, Steven.dcm has already been uploaded for testing purposes, but users can try uploading the remaining DICOMs. Also, the dropdown for past scans will only show uploaded scans for the currently selected patient. The database is not publicly accessible, so feel free to reach out to our team if you would like the demo to be reset.
-
Run the front end test cases.
python3 test_index.py
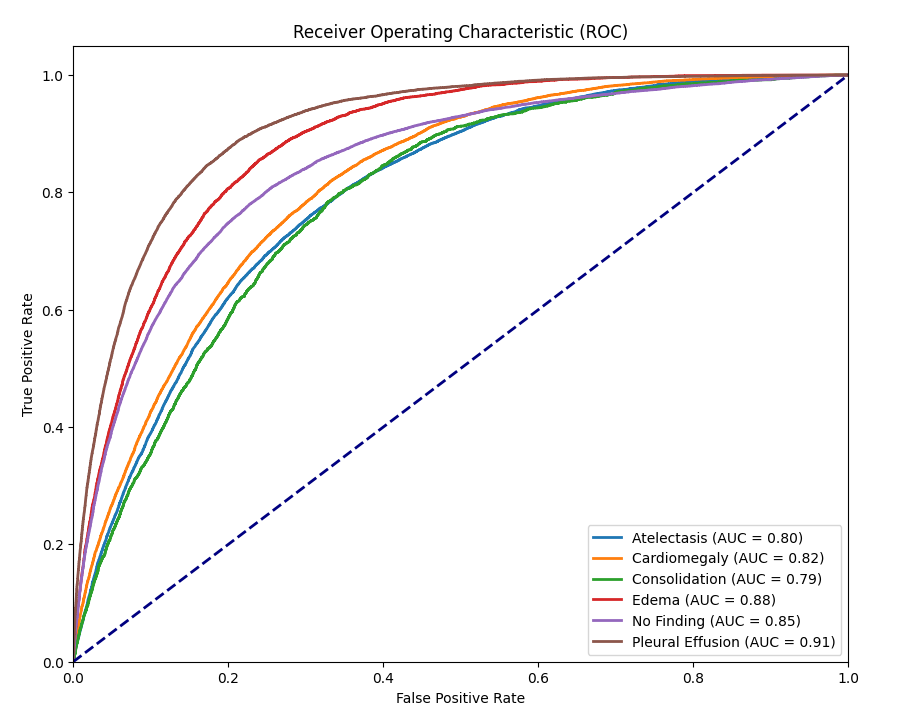
| Atelectasis | Cardiomegaly | Consolidation | Edema | No Finding | Pleural Effusion | |
|---|---|---|---|---|---|---|
AUC |
0.7969 | 0.8154 | 0.7893 | 0.8831 | 0.8483 | 0.9110 |
Best Threshold |
0.1711 | 0.1580 | 0.0385 | 0.0980 | 0.3500 | 0.1623 |
Precision |
0.3395 | 0.3372 | 0.0837 | 0.2730 | 0.6581 | 0.5287 |
Recall |
0.7731 | 0.8190 | 0.7979 | 0.8564 | 0.8095 | 0.8859 |
F1 Score |
0.4718 | 0.4777 | 0.1515 | 0.4140 | 0.7260 | 0.6622 |
| Finding | AUC Defined in P0 | Validation AUC | Final Testing AUC |
|---|---|---|---|
Atelectasis |
0.80 | 0.8029 | 0.7969 |
Cardiomegaly |
0.85 | 0.8130 | 0.8154 |
Consolidation |
0.85 | 0.7951 | 0.7893 |
Edema |
0.85 | 0.8861 | 0.8831 |
No Finding |
0.85 | 0.8487 | 0.8483 |
Pleural Effusion |
0.92 | 0.9175 | 0.9110 |
Dataset: MIMIC-CXR-JPG
The dataset contains 10 folders p10-p19 and here is the data split:
Training (70%): Folders 10-16
Validation (10%): Folder 17
Testing (20%): Folders 18 and 19
-
PyTorch Grad Cam
https://github.com/jacobgil/pytorch-grad-cam?tab=MIT-1-ov-file
-
TorchXRayVision
https://github.com/mlmed/torchxrayvision?tab=Apache-2.0-1-ov-file
-
MIMIC Dataset Resources
https://doi.org/10.13026/jsn5-t979
https://physionet.org/content/mimic-cxr/2.0.0/
Johnson, A., Lungren, M., Peng, Y., Lu, Z., Mark, R., Berkowitz, S., & Horng, S. (2024). MIMIC-CXR-JPG - chest radiographs with structured labels (version 2.1.0). PhysioNet.
Johnson, A. E., Pollard, T. J., Greenbaum, N. R., Lungren, M. P., Deng, C. Y., Peng, Y., ... & Horng, S. (2019). MIMIC-CXR-JPG, a large publicly available database of labeled chest radiographs. arXiv preprint arXiv:1901.07042.
Goldberger, A., Amaral, L., Glass, L., Hausdorff, J., Ivanov, P. C., Mark, R., ... & Stanley, H. E. (2000). PhysioBank, PhysioToolkit, and PhysioNet: Components of a new research resource for complex physiologic signals. Circulation [Online]. 101 (23), pp. e215–e220.