This repository contains scripts and tools that run code for the paper titled ‘ModusGraph: Automated 3D and 4D Mesh Model Reconstruction from cine CMR with Improved Accuracy and Efficiency’. The sole purpose of this repository is to provide a reference for the paper. Please note that the code is written in Python 3.9.16 and Pytorch 1.12.1, and is not optimized for efficiency or guaranteed to be bug-free.
To gain a better understanding of the idea behind the network design and parameter settings, we invite you to watch the following brief presentation using your MICCAI 2023 access.
The code is tested on Ubuntu 18.04.6 LTS. To install the code, first clone the repository:
$ git clone https://github.com/MalikTeng/ModusGraph
Then install the conda environment:
$ conda env create -f environment.yml
Then activate the environment:
$ conda activate modusgraph
The data used in the paper is not included in this repository. The data is available upon request, and details provided in the paper. The data is organized in the following structure:
data
├── imagesTr
│ ├── 1.nii.gz
│ ├── 2.nii.gz
│ ├── ...
│ └── 100.nii.gz
├── labelsTr
│ ├── 1.nii.gz
│ ├── 2.nii.gz
│ ├── ...
│ └── 100.nii.gz
Template meshes were needed for runing the code. The template meshes are included for reference. The template meshes are organized in the following structure:
template
|── scotheart
| ├── 1.obj
| ├── 2.obj
| ├── ...
| └── 6.obj
|── cap
| ├── 1.obj
Data preprocessing is a must so that images, segmentations, and template meshes are in the same space. While ways to conduct such a preprocessing are not included in this script, the result can be varified by the script 'test_XXXX.py'.
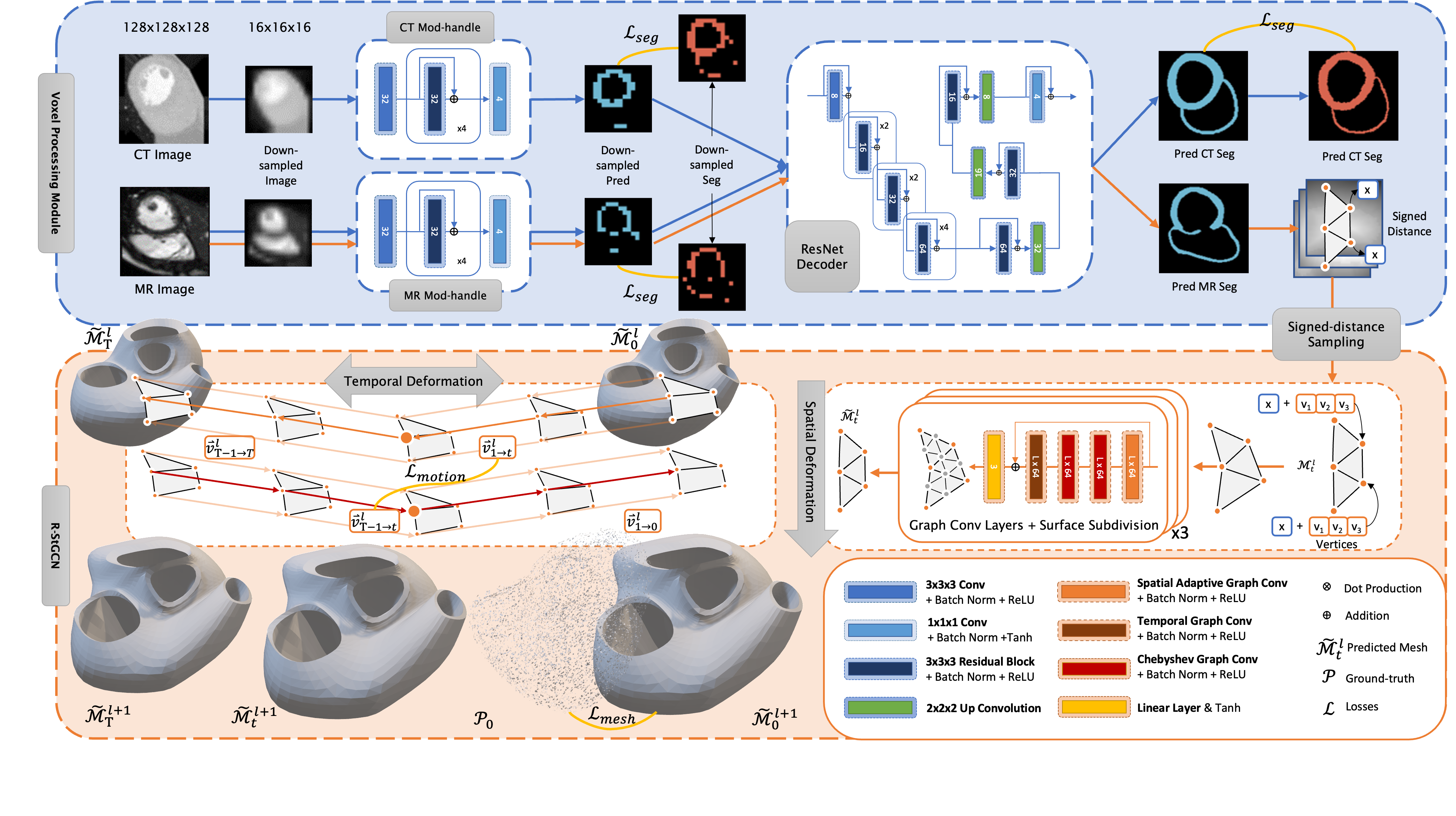
Detail structure of ModusGraph can be found in the paper.
The training process contains two stages:
- Pre-training: train the Voxel Processing Module, including CT modality handel and ResNet decoder.
- Training: train the R-StGCN module.
The training process contains two stages:
- Pre-training: train the Voxel Processing Module and the R-StGCN module with CT data.
- Training: fine-tuning the whole ModusGraph networks with cine CMR data.
Parameters of the network is customizable but not recommended.
We recommand using Weights and Bias for monitoring the training process. Following snapshot shows the training process of the two training stages.
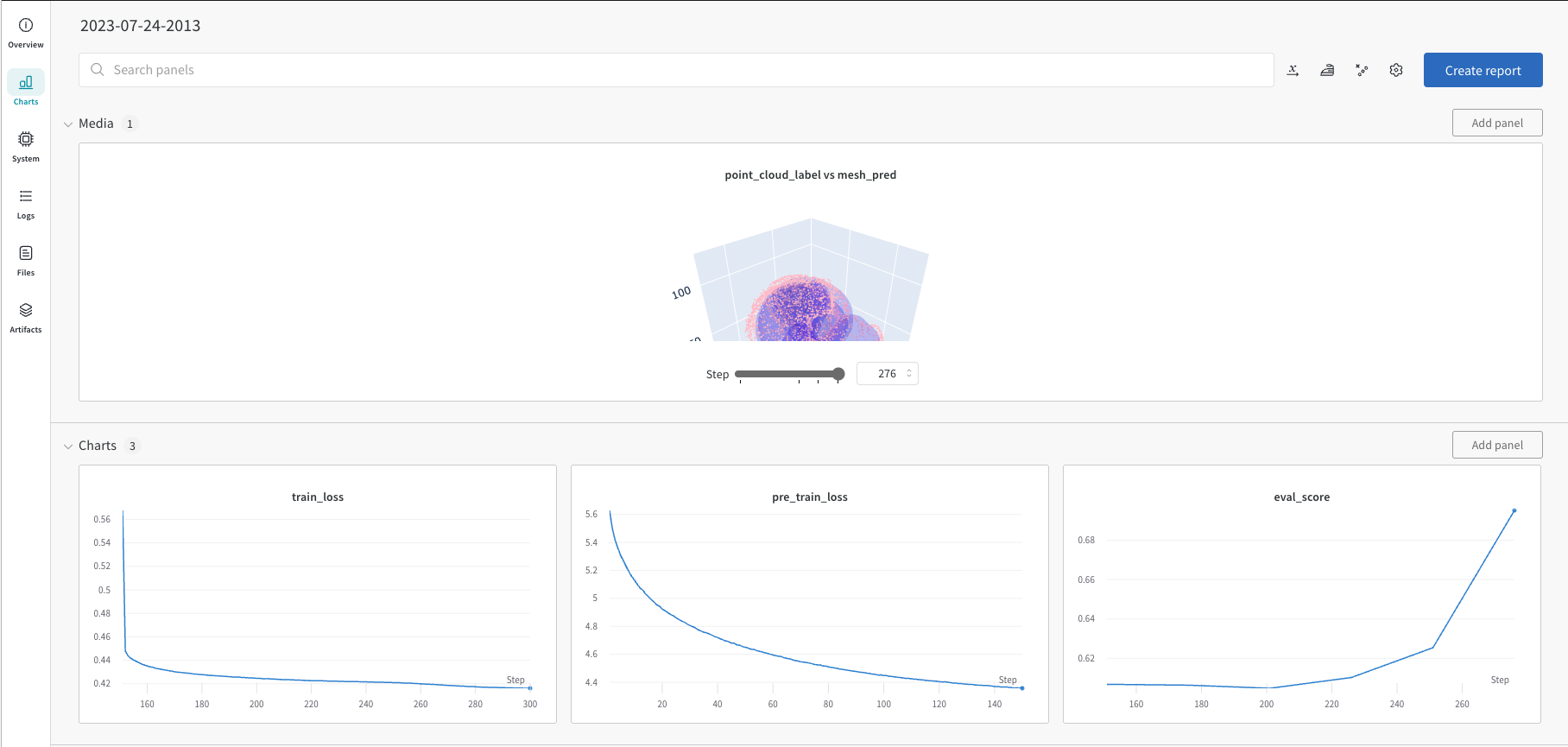
Running the whole training process is straightforward, just use commandline tool. See details of adjustable parameters in the script.
$ python main-whole_heart_meshing.py \
--image_dir /path/to/image \
--label_dir /path/to/label \
--log_dir /path/to/log_dir \
--out_dir /path/to/out_dir \
--max_epochs 500 \
--delay_epochs 250 \
--val_interval 50 \
--crop_window_size 128 128 128 \
--batch_size 24 \
--point_limit 53_500 \
--cache_rate 1.0
Running the dynamic meshing process is very similar except arguments need for both CT and MR images and labels. See details in the script.
$ python main-dynamic_meshing.py \
--ct_image_dir /path/to/image_ct \
--ct_label_dir /path/to/label_ct \
--mr_image_dir /path/to/image_mr \
--mr_label_dir /path/to/label_mr \
--log_dir /path/to/log_dir \
--out_dir /path/to/out_dir \
--max_epochs 500 \
--delay_epochs 250 \
--val_interval 50 \
--crop_window_size 128 128 128 \
--batch_size 24 \
--point_limit 53_500 \
--cache_rate 1.0