Contribution to the SEG.A Challenge (MICCAI 2023) by Marek Wodzinski.
The repository presents the contribution to the SEG.A Challenge that scored:
- 1st place in the clinical evaluation
- 4th place in the evaluation based on quantiative metrics
- 3rd place in the volumetric meshing quality
The challenge website: Link
The repository provides the full source code used to train / test the proposed solution.
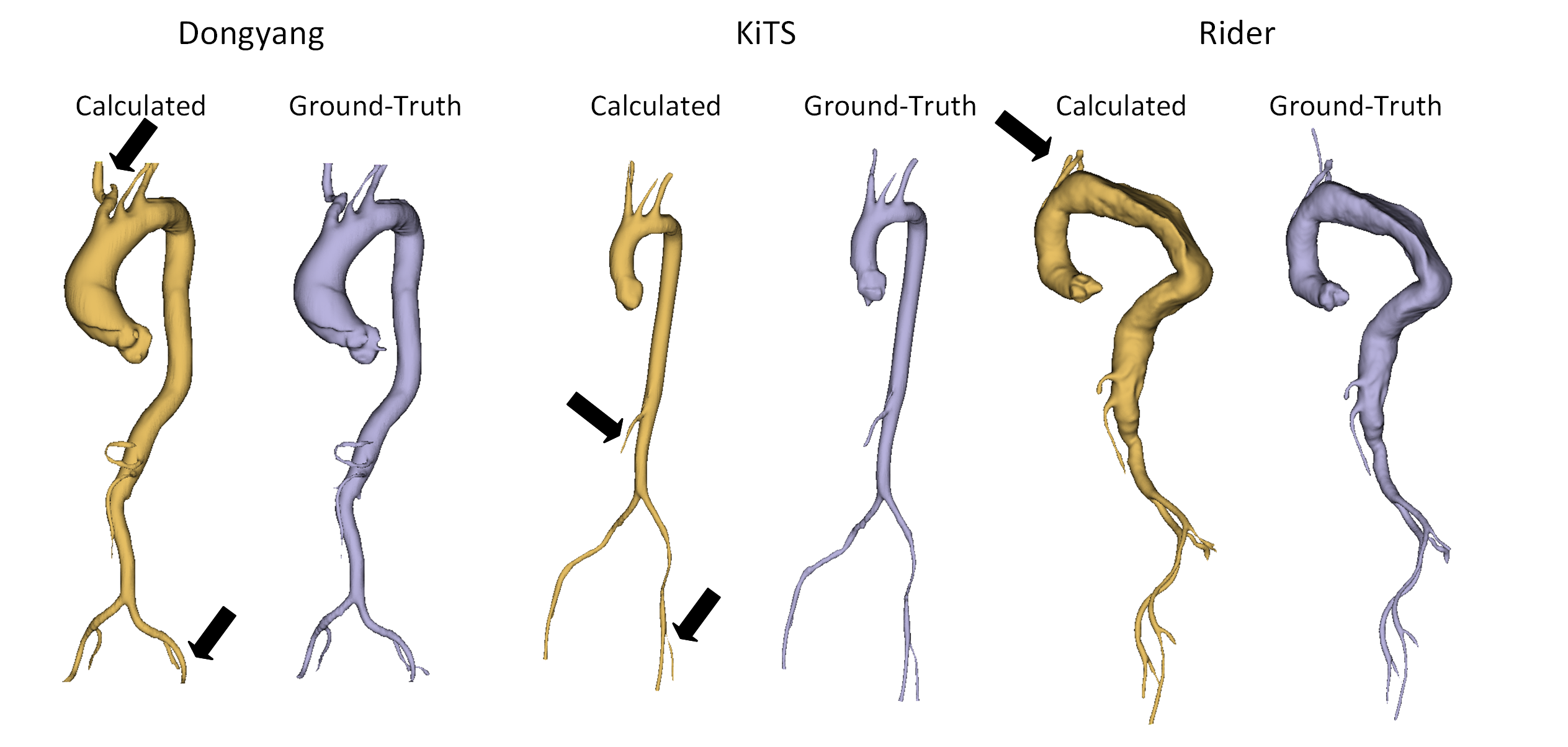
The exemplary qualitative results obtained by the method and compared to the ground-truth are visible below:
Exemplary cases from each medical center from the cross-validation dataset:
And finally, the quantitative comparison to other challenge participants is as follows:
Only the final experiment is available in the repository (the one used for the final Docker submission).
- In order to reproduce the experiment you should:
- Download the SEG.A dataset Link
- Update the hpc_paths.py and paths.py files.
- Run the parse_sega.py
- Run the run_aug_sega.py
- Run the training using run_segmentation_trainer.py
- And finally use the trained model for inference using inference.py
The network was trained using HPC infrastructure (PLGRID). Therefore the .slurm scripts are omitted for clarity.
In case you are interested in just running the code:
- If you want to run the model locally - the final model is available upon reasonable request.
- You can also apply for an access to the algorithm directly using the Grand-Challenge platform: Link. Access will be provided without restrictions.
If you found the source code useful, please cite:
- The SEG.A Challenge Paper - Will Be There Soon
- The paper presenting the proposed solution:
We gratefully acknowledge Polish HPC infrastructure PLGrid support within computational grant no. PLG/2023/016239.