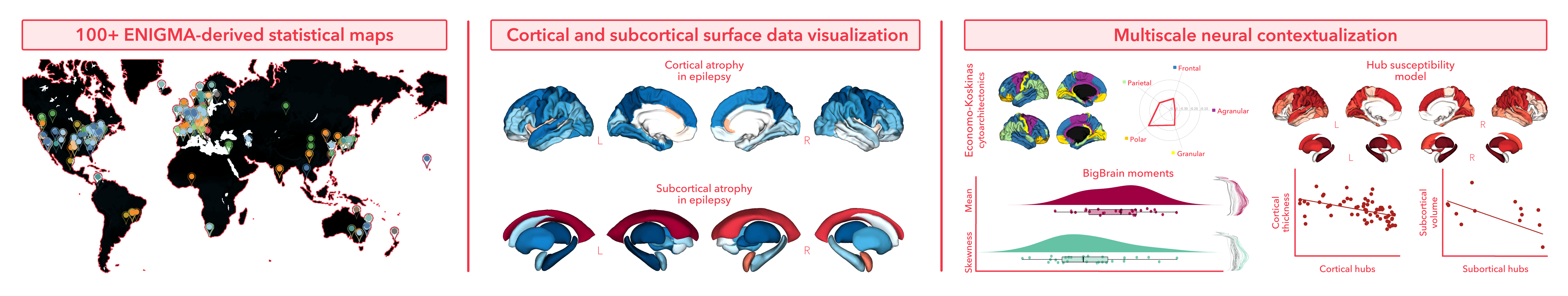
The ENIGMA TOOLBOX is an open source repository for (i) accessing 100+ ENIGMA-derived statistical maps, (ii) visualizing and manipulating cortical and subcortical surface data, and (iii) contextualizing neuroimaging findings at the microscale (using postmortem gene expression and cytoarchitecture) and macroscale (using structural and functional connectome data).
Check out our expandable online documentation at https://enigma-toolbox.readthedocs.io to learn how to:
To install the Toolbox in Python, run the following in your terminal:
git clone https://github.com/MICA-MNI/ENIGMA.git
cd ENIGMA
python setup.py installTo install the Toolbox in Matlab, run the following in your terminal:
git clone https://github.com/MICA-MNI/ENIGMA.gitAnd then simply run the following in Matlab:
addpath(genpath('/path/to/ENIGMA/matlab/'))Please acknowledge this work using the citation below:
Larivière S, Paquola C, Park BY, Royer J, Wang Y, Benkarim O, Vos de Wael R, Valk SL, Thomopoulos SI, Kirschner M, ENIGMA Consortium, Sisodiya S, McDonald C, Thompson PM, Bernhardt BC (2020). The ENIGMA Toolbox: Cross-disorder integration and multiscale neural contextualization of multisite neuroimaging datasets. bioRxiv, https://doi.org/10.1101/2020.12.21.423838.