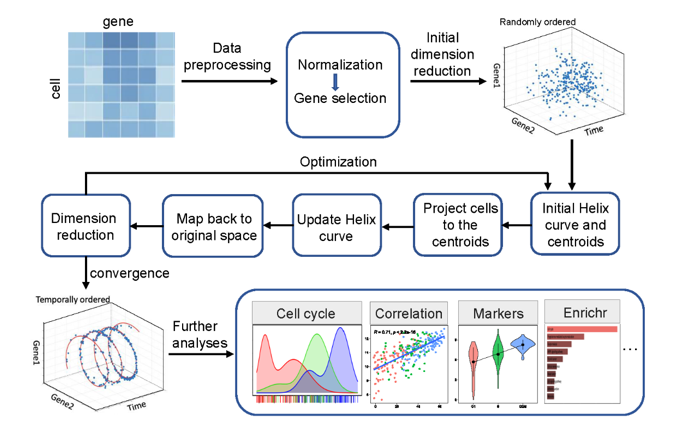
CCPE is a cell cycle pseudotime estimation method characterizing cell cycle timing from single-cell RNA-seq data. CCPE maps high-dimensional scRNA-seq data onto a helix in three-dimensional space, where 2D space is used to capture the cycle information in scRNA-seq data, and one dimension to predict the chronological orders of cells along the cycle, which we called cell cycle pseudotime. ScRNA-seq data is repeatedly transformed from high dimensional to low dimensional and then mapped back to high dimensional. At the same time, CCPE iteratively optimizes the discriminative dimensionality reduction via learning a helix until convergence (Figure 1). CCPE is applied to several downstream analyses and applications to demonstrate its ability to accurately estimate the cell cycle pseudotime and stages.
Our paper was published in Nucleic Acids Research, available at Jiajia Liu, Mengyuan Yang, Weiling Zhao, Xiaobo Zhou, CCPE: cell cycle pseudotime estimation for single cell RNA-seq data, Nucleic Acids Research, 2021
drtoolbox was downloaded from https://lvdmaaten.github.io/drtoolbox/
wget https://github.com/LiuJJ0327/CCPE/archive/refs/heads/main.zip
unzip CCPE-main.zip
cd CCPE-main/
matlab run.mRscript pseudotime_to_label.r The reproduction of the applications and analyses in our paper can be available at reproduction/
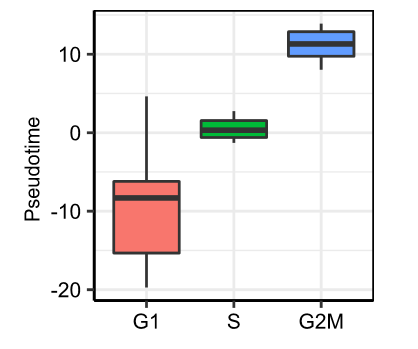
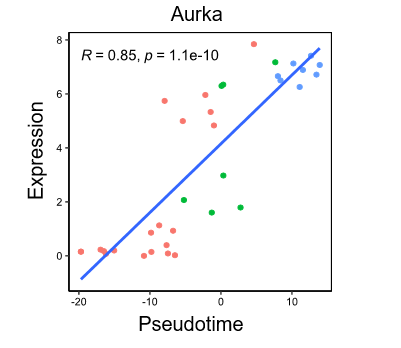
Rscript reproduction/pseudotime_analysis/4_analysis_R/plot_pseudotime.rRscript reproduction/pseudotime_analysis/4_analysis_R/correlation.rlibrary(Seurat)
mesc<-read.table("./pseudotime_analysis/1_mesc_Quartz_data/mESC_Quartz_preprocessed.txt",header=T,row.names = 1)
marrow <- CreateSeuratObject(raw.data = mesc)
marrow@ident<-factor(marrow@ident,levels = c("G1","S","G2M"))
RidgePlot(object =marrow, features = 'Aurka')