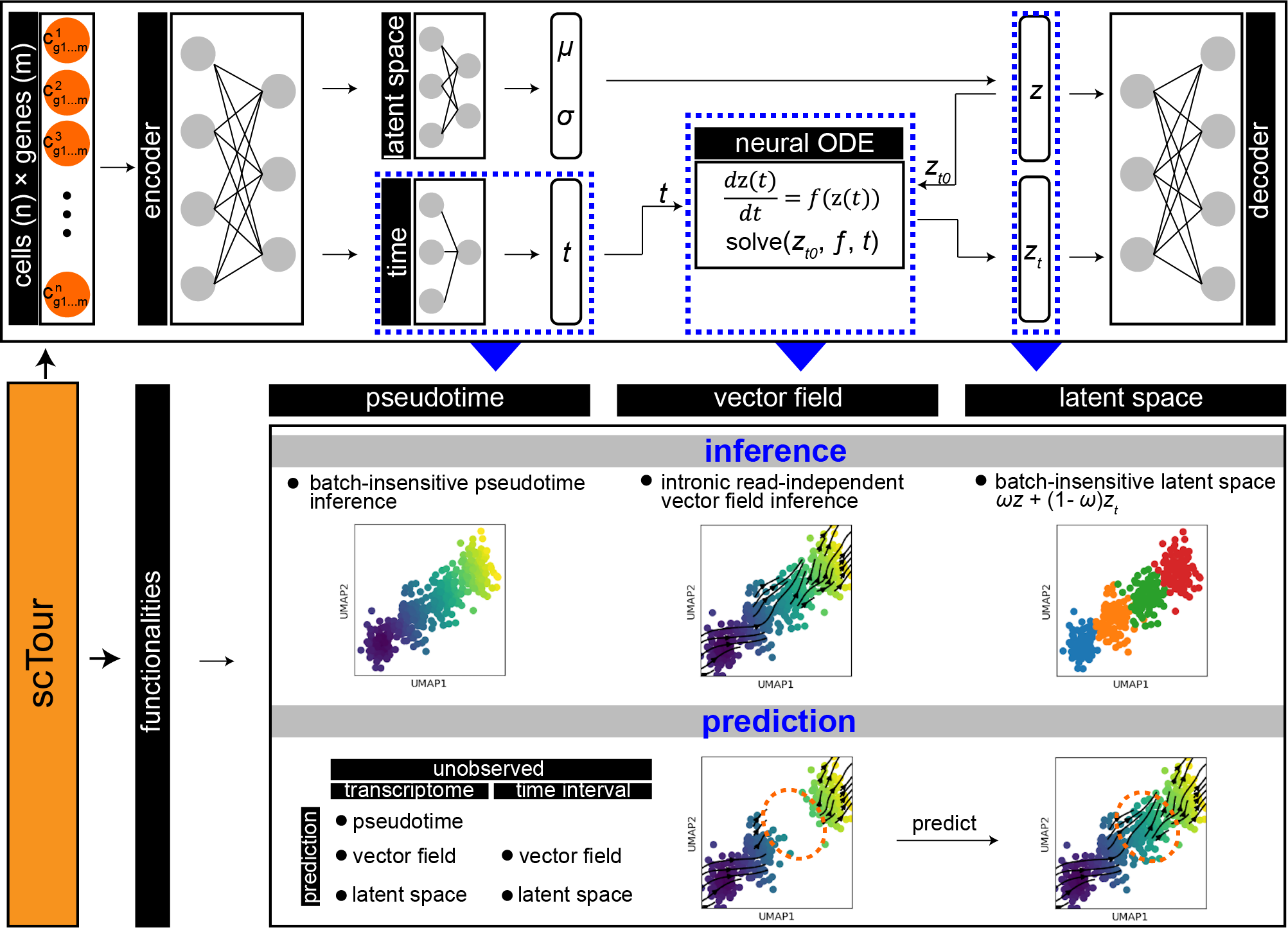
scTour is an innovative and comprehensive method for dissecting cellular dynamics by analysing datasets derived from single-cell genomics.
It provides a unifying framework to depict the full picture of developmental processes from multiple angles including the developmental pseudotime, vector field and latent space.
It further generalises these functionalities to a multi-task architecture for within-dataset inference and cross-dataset prediction of cellular dynamics in a batch-insensitive manner.
- cell pseudotime estimation with no need for specifying starting cells.
- transcriptomic vector field inference with no discrimination between spliced and unspliced mRNAs.
- latent space mapping by combining intrinsic transcriptomic structure with extrinsic pseudotime ordering.
- model-based prediction of pseudotime, vector field, and latent space for query cells/datasets/time intervals.
- insensitive to batch effects; robust to cell subsampling; scalable to large datasets.
pip install sctourconda install -c conda-forge sctourFull documentation can be found here.