Implementation of noise reduction algorithms applying to electron density maps. This code is part of master's qualification diploma. The purpose is to show how good BM4D (3d case of BM3D (Block-matching and 3D filtering)) works with ed-maps.
Example of current implementation BM4D denoise algorithm applying to EMD-2984 map visualized through miew visualizer.


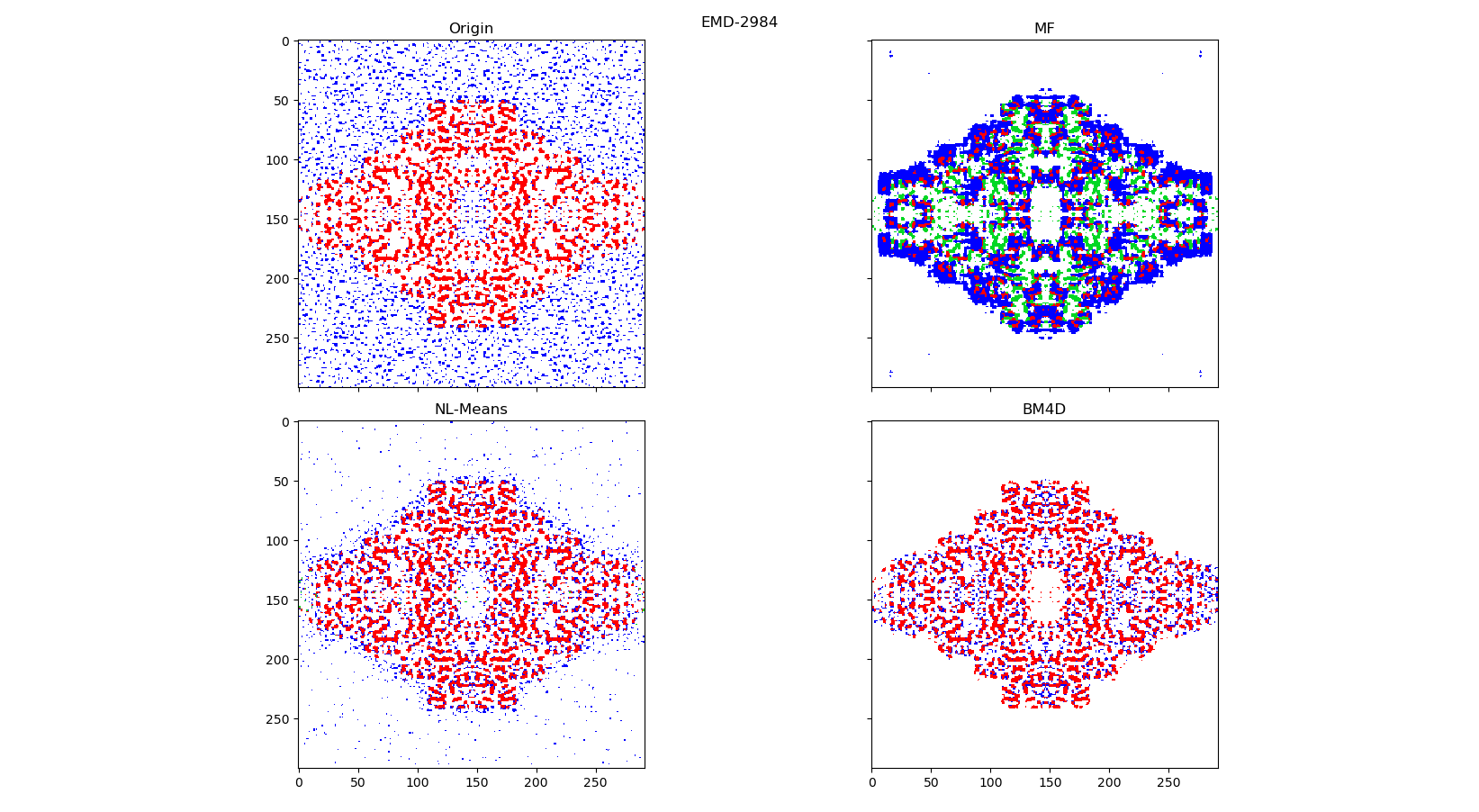
The left picture is the origin EMD-2984 map and the right picture is denoised by current BM4D algorithm map.
- Open any molecular viewer in the first tab, suggested miew.
- Load origin EMD-2984 map (in miew: open menu, choose load, load from file, chose EMD-2984_origin.ccp4 from ED-maps-denoised-results archive or from emdb site).
- Open molecular viewer in the second tab,
- Load bm4d denoised map (in miew: open menu, choose load, load from file, choose EMD-2984_bm4d.ccp4.
- Enable representing several molecules in the viewer, if not so by default (in miew: Open terminal (arrow in upper-left corner), write in terminal “set use.multiFile 1”)
- Load “5a1a” PDB (it is PDB model of EMD-2984 map), (in miew: Open menu, choose load, write “5a1a”)
- Additionally, for better view in miew: Open “Display mode” (upper-right) corner, choose balls mode
Electron density maps can be viewed as single-channel 3D images indicating the density of a substance at each point (cell).
The vast majority of cells in molecular density maps are empty, that is, they are filled with noise.
If one visualize all the cells indiscriminately, one gets an almost uniform box in which nothing can be disassembled.
For the most part, the values of cells with a signal are higher than those of cells with noise.
Therefore, in order to visualize a signal, some threshold number is usually chosen (default: expected value + 3 sigma)
and cells whose value is greater than the threshold number are considered as signal and are visualized, the rest are considered as noise and are not displayed.
However, it is not always easy to find such a threshold number, moreover, the values of some noisy cells exceed the threshold,
and such cells are visualized.
Proposed solution:
A model is proposed in which the ed-map is considered as a 3D image with white Gaussian noise superimposed.
Then supposed to apply noise reduction algorithms to these maps.
- Median filter with 2d and 3d window.
- Non-local means with 2d and 3d window.
- Block-matching and 3D filtering (BM3D) and generalization to 3d input picture - BM4D.
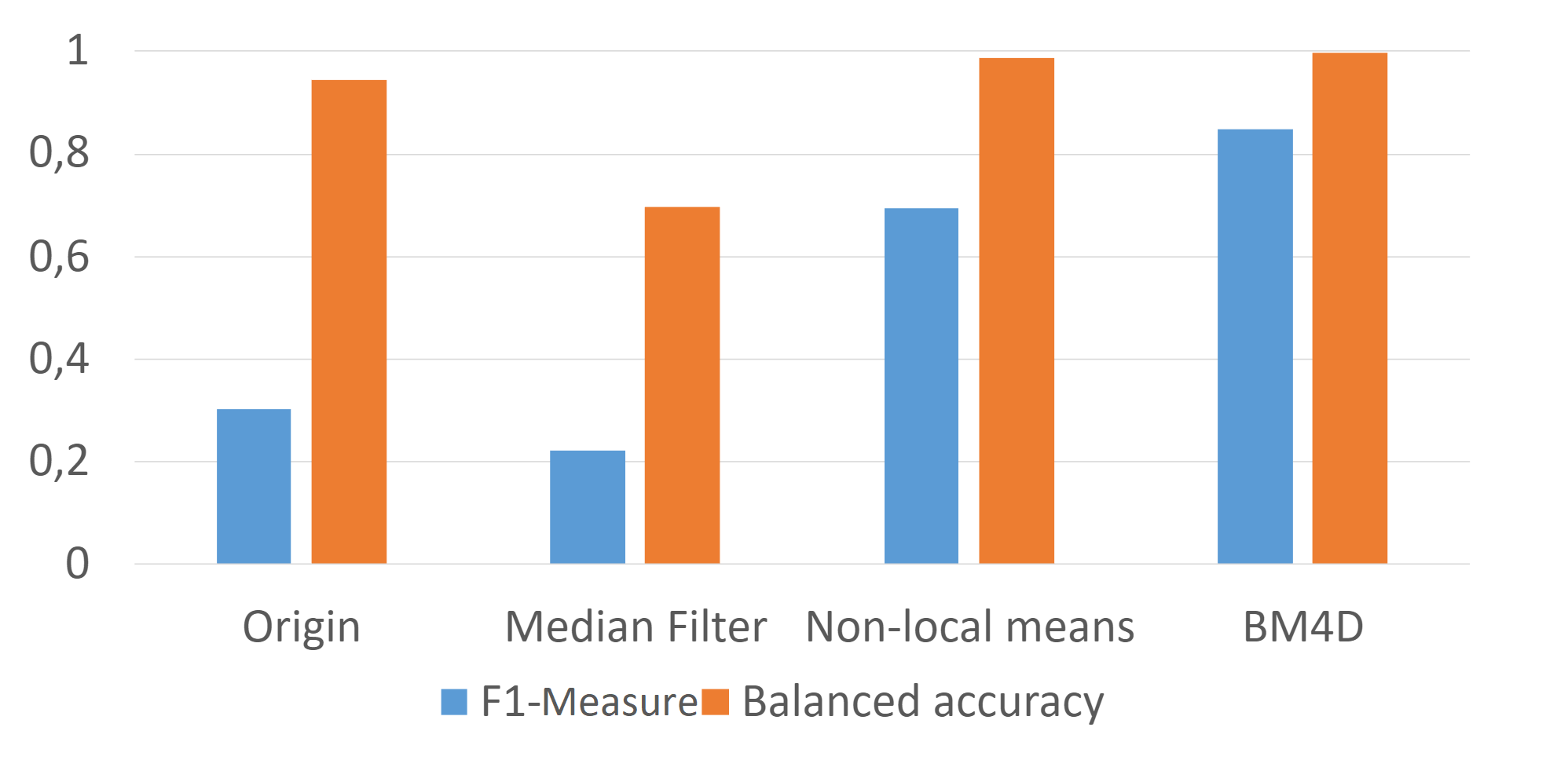
For non-local means and BM3D-BM4D applying an approach to matching a random subset (not all, but a sufficient number) of matching blocks. This approach is based on Monte Carlo non local means: Random sampling for large-scale image filtering article. The implementation of BM3D-BM4D is not entirely done (omitted second step with filtering in the spectral area). But just the first step already demonstrates satisfactory results.
Code supported ccp4 (.map) and dsn6 formats of electron density maps. So there are
ccp4 and
dsn6 parsers accordingly.
Class MolRepresentation consists of:
- 3-d NumPy array which is the content of ed-map
- header of ed-map (some additional meta information)
- statistics information (required for normalization)
- Example of reading ccp4 ed-map:
from scripts.ed_parser import read
from scripts.molecule import MolRepresentation
filePath = 'EMD-6479.ccp4'
ed: MolRepresentation = read(filePath)- Example of running denoising algorithm:
import denoise_methods.BM4D as BM4D
threshold = 4.85
ed.re_normalize() # normalization is obligatory!
denoiser = BM4D.BM4D(ed.values)
denoise_data = denoiser.execute_3d(threshold)- Example of writing denoised data to file
Denoise algorithms work only with 3d data (ed.values). So to save new data the easiest way is to use updated header information from the original map:
from scripts.ccp4_parser import to_ccp4_file
import numpy as np
ed.update_from_values(denoise_data)
ed.header.mean = np.mean(ed.buffer)
ed.header.stddev = np.std(ed.buffer)
ed.header.min = np.min(ed.buffer)
ed.header.max = np.max(ed.buffer)
ed.header.fields["amean"] = ed.header.mean
ed.header.fields["amax"] = ed.header.max
ed.header.fields["amin"] = ed.header.min
ed.header.fields["sd"] = ed.header.stddev
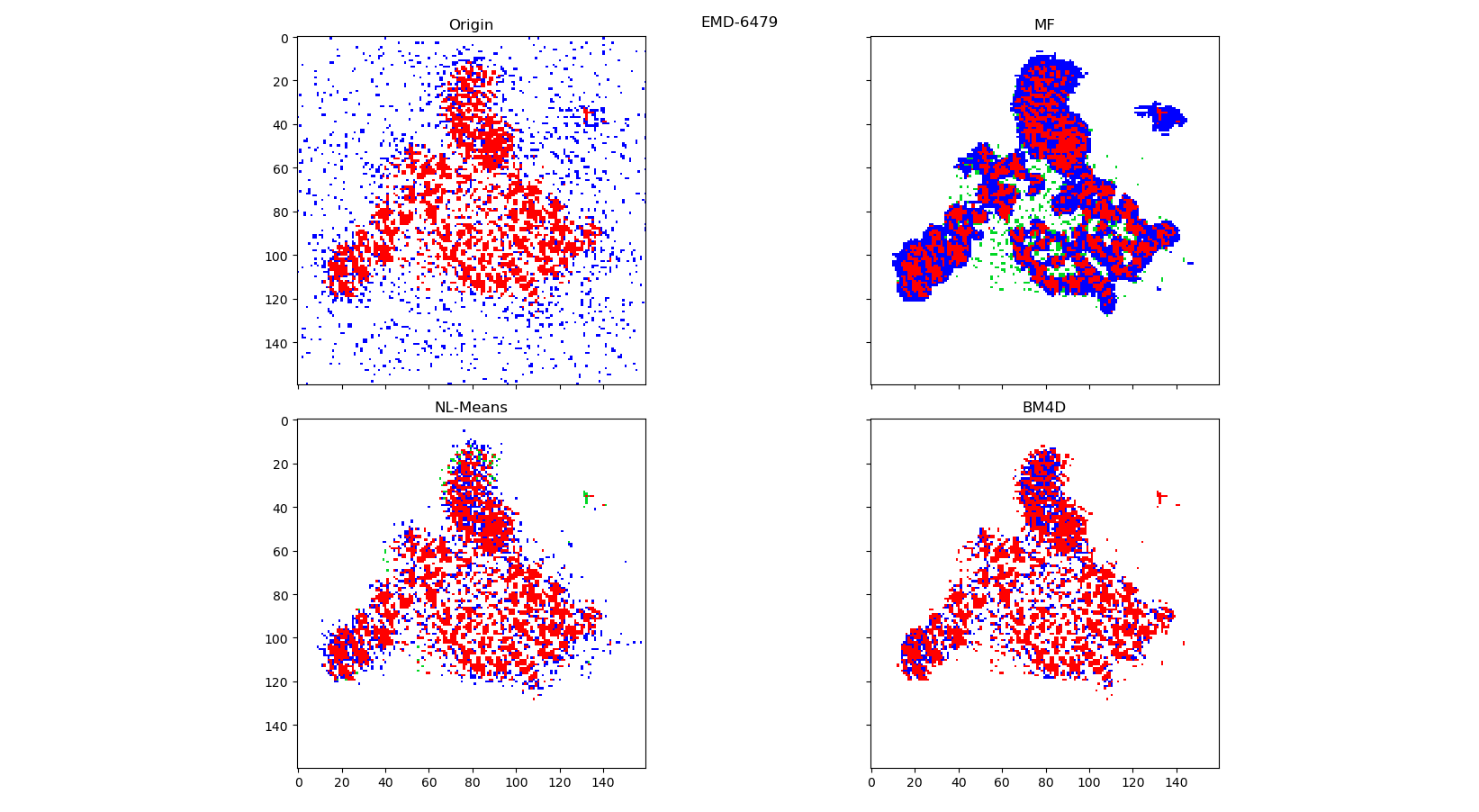
to_ccp4_file(ed, 'bm4d')To test denoise algorithms it is been taken several ed-maps with varying degrees of noise: 4NRE, EMD-6479, EMD-2984.
Original maps and best results for each method (2d and 3d) are laid in ED-maps-denoised-results archive.
Red: True positive
Blue: False positive
White: True negative
Green: False negative