Identification and quantification of alternative polyadenylation sites from single-cell RNA-seq data
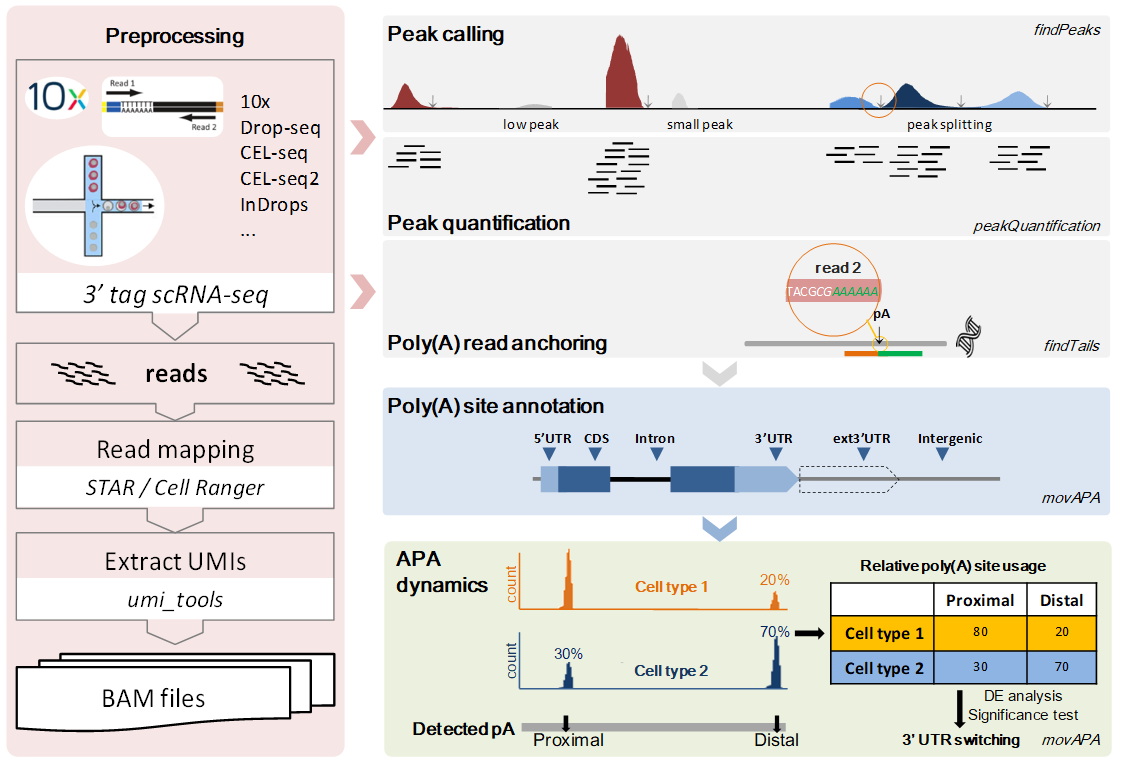
Alternative polyadenylation (APA) has been indicated to play an important role in regulating mRNA stability, translation and localization. Diverse scRNA-seq protocols, such as Drop-seq, CEL-seq, and 10x Genomics, utilizing 3' selection/enrichment in library construction, provide opportunities to extend bioinformatic analysis for studying APA at single cell resolution. We proposed a tool called scAPAtrap for the identification and quantification of APA sites in each individual cells by leveraging the resolution and huge abundance of scRNA-seq data generated by various 3' tag-based protocols. scAPAtrap incorporates peak identification and poly(A) read anchoring, which is capable of identifying and pinpointing poly(A) sites even for those with low read coverage. scAPAtrap can also quantify expression levels of all identified APA sites, considering duplicates resulted from both IVT and PCR cycles.
scAPAtrap mainly consists of six modules. (1) Raw scRNA-seq datasets from 3’ tag-based protocols (e.g., 10x, CEL-seq) were preprocessed for mapping and extracting UMIs. (2) Without using any genome annotation, potential peaks of the whole genome were detected and wide peaks were iteratively splitted into smaller ones. (3) Identified peaks are quantified by counting effective reads in the peaks. (4) Reads with A/T streches were extracted to determine precise locations of poly(A) sites. (5) Poly(A) sites in both genomic and intergenic regions were annotated with rich information according to the latest genome annotation. (6) Differentially expressed poly(A) sites and 3′ UTR lengthening/shortening events were detected to profile APA dynamics among cell types.
The above tools can be installed using conda.
conda install samtools -c bioconda
conda install subread -c bioconda
conda install umi_tools -c bioconda
conda install star -c bioconda
Please install the following R packages:
GenomicRanges GenomicAlignments dplyr derfinder regionR
Three main datasets used in this study:
- Mouse spermatogenesis (GSE104556)
- Arabidopsis roots (GSE123013)
- Mouse intestinal organoids (GSE62270)
Lists of poly(A) sites with full genome annotation identified by scAPAtrap were placed in the Result folder.
install.packages('devtools')
devtools::install_github("BMILAB/scAPAtrap",build_vignettes = TRUE)
Note, due to the problem of the devtools version, there may be no build_vignettes parameter, you can use the following command to download scAPAtrap.
devtools::install_github("BMILAB/scAPAtrap", build_opts = c("--no-resave-data", "--no-manual"))
In this case study, we investigated the application of scAPAtrap on identifying and quantifying poly(A) sites from mouse spermatogenesis scRNA-seq data. Please refer to the vignette (scAPAtrap.html) for full details.
## You can also browse the vignette using the following command on the R console.
browseVignettes('scAPAtrap')
Here we adopted the mouse sperm scRNA-seq dataset to evaluate the performance of scAPAtrap and compared the results with other two tools, scAPA (Shulman et al, 2019) and Sierra (Patrick, et al., 2020). We have used scAPAtrap, scAPA, and Sierra to identify poly(A) sites from the mouse sperm scRNA-seq data, respectively. The identified poly(A) sites stored in Rdata files can be downloaded here. Please refer to the vignette (scAPAtrap_compare.html) for full details.
We analyzed dynamic APA usage during sperm cell differentiation based on poly(A) sites identified by scAPAtrap. Please refer to the vignette (scAPAtrap_DE.html) for full details.
If you are using scAPAtrap, please cite: Xiaohui Wu*, Tao Liu, Congting Ye, Wenbin Ye, Guoli Ji: scAPAtrap: identification and quantification of alternative polyadenylation sites from single-cell RNA-seq data, Briefings in Bioinformatics, 2020.