Deep supervision spatial Feature Fusion convolutional network (DS-SFFNet) is an end-to-end liver and liver tumor segmentation neural network, which based on deep supervision mechnaism and residual attention mechnaism.
This work used miriambellver's work and modified it to achieve our purpose.
In the training phase, we first train liver segmentation network to get the coarse liver segmentation results. The liver ROI of CT images are located according to the liver segmentation results. Secondly, we use liver ROI to train liver tumor segmentation network to get the coarse liver tumor segmentation results.
In the testing phase, we first get the accurate liver segmentation results. Then the DS-SFFNet can generate the coarse liver tumor segmentation results. To avoid over-segmentation in the liver tumor region, we remove the area out of liver by using the liver mask. In the end, a 3D fully connected conditional random fields(CRFs) is utilized to refine the final liver tumor segmentation results.
In DS-SFFNet, we add rich skip-connections to recover detailed spatial features based on miriambellver's network architecture. We also propose a novel feature fusion block to fuse detailed spatial feartures and high-level semantic feature.
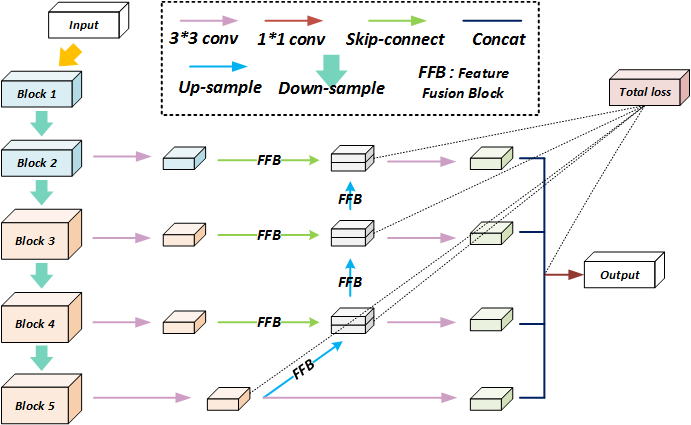
The details of DS-SFFNet architecture. At the down-sample part, there are 13 convolutional layers and 4 max pooling layers. The network is divided into different convolutional blocks according to the resolution. And the side-output layers are connected at the end of each block to supervise the training process and compute the final loss. At the up-sample part, we use the bilinear interpolation to recovery the size of feature maps and introduce the FFB (the structure of FFB is in section B) to reserve more spatial features. Therefore, the feature maps from skip-connections and up-sampling layers can properly stack together.
we design a feature fusion block to recover spatial features at decoder part and fuse feature maps from different resolution.
We use the contrast-enhanced abdominal CT scans of MICCAI 2017 Liver Tumor Segmentation (LiTS) challenge dataset (https://competitions.codalab.org/competitions/17094) to test our model. Our liver segmentation results get 0.955 Dice global score, 0.937 Dice per volume score and 0.106 volume overlap error score. The tumor detected precision has improved and the score is 0.369. The Dice global score is 0.746 and the Dice per volume score is 0.592.
@misc{1711.11069,
Author = {Miriam Bellver and Kevis-Kokitsi Maninis and Jordi Pont-Tuset and Xavier Giro-i-Nieto and Jordi Torres and Luc Van Gool},
Title = {Detection-aided liver lesion segmentation using deep learning},
Year = {2017},
Eprint = {arXiv:1711.11069},
}
3D CRFs : https://github.com/mbickel/DenseInferenceWrapper