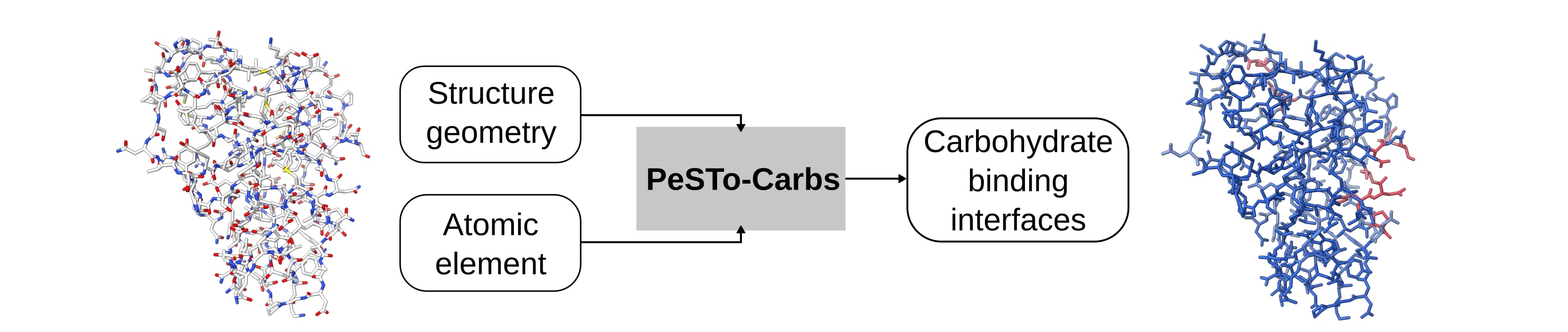
PeSTo-Carbs is an extension of PeSTo, trained to predict protein-carbohydrate and protein-cyclodextrin binding interfaces. We provide two models for PeSTo-Carbs: a general model PS-G (ps-s.pt) for a wide range of carbohydrates, their derivatives and cyclodextrins and a specific PS-S (ps-s.pt) trained on specific important carbohydrate monomers. The method is available on our webserver at pesto.epfl.ch.
Clone this repo or download the source code. To install dependencies, run
pip install -r requirements.txt
The apply_model.py can be used to make predictions on PDB files. In the file model_path can be modified to specify ps-g.pt or ps-s.pt and data_path can be set to file paths for PDB files. For each PDB, two files will be generated, <pdbid>_i0.pdb for protein-carbohydrate prediction and <pdbid>_i1.pdb for protein-cyclodextrin prediction. The predicted values are stored in the b-factor column. This can be visualized in PyMOL using:
spectrum b, blue_white_red, all, 0, 1
Or in ChimeraX using:
color bfactor palette "#2B59C3:#D1D1D1:#D7263D" range 0,1
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License.
Bibekar, P., Krapp, L., Dal Peraro, M. et al. PeSTo-Carbs: Geometric Deep Learning for Prediction of Protein−Carbohydrate Binding Interfaces. J. Chem. Theory Comput., 2024. DOI: 10.1021/acs.jctc.3c01028.