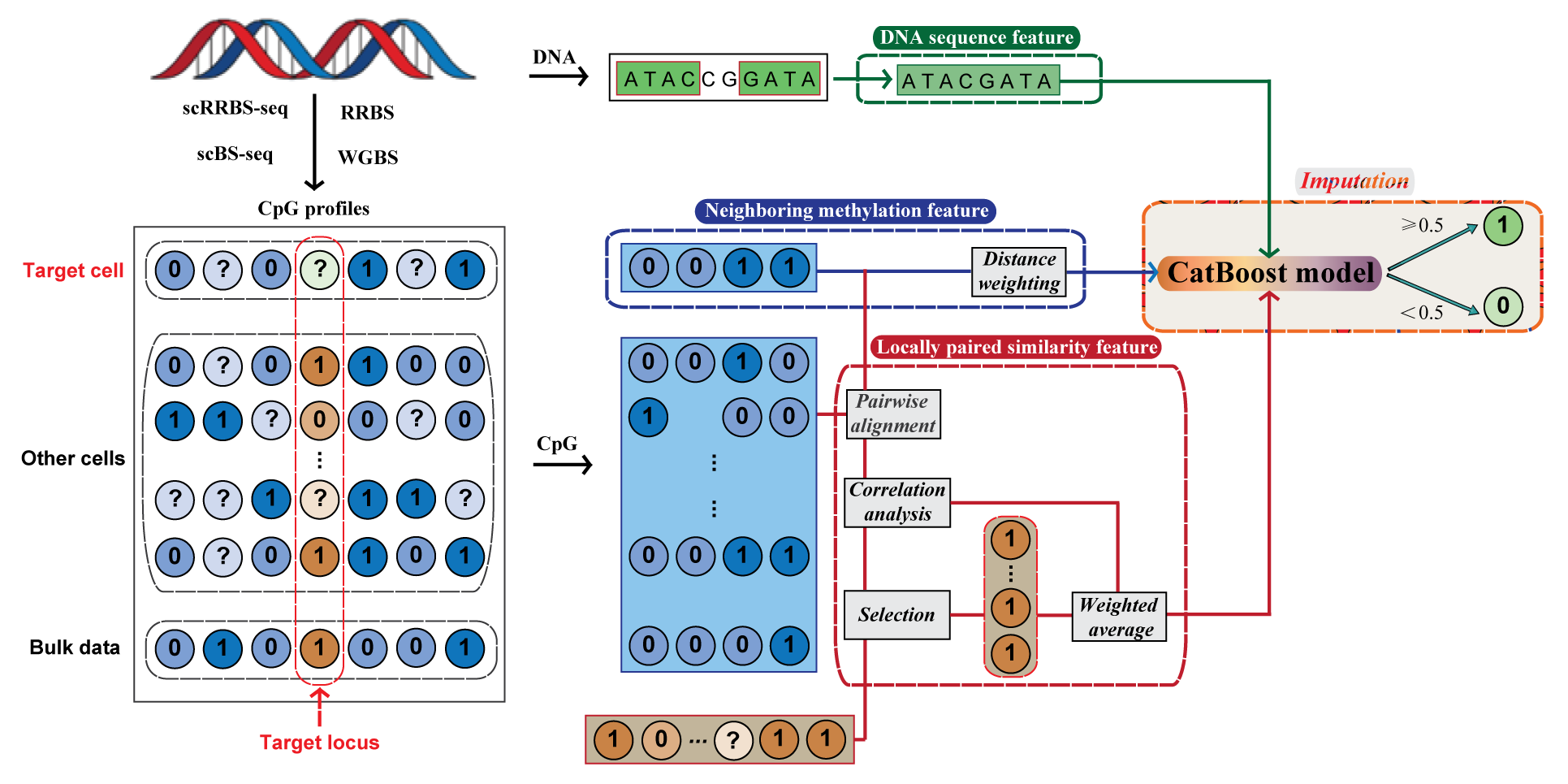
CaMelia (CAtboost-based method for predicting MEthyLatIon stAtes), a computational imputation method based on the CatBoost gradient boosting model for predicting single-cell methylation states to address the sparsity of the data. CaMelia imputed the missing methylation states in a cell by borrowing information of the same locus in other cells, based on the locally paired similarity of methylation patterns around the target locus between cells or between cell and bulk data. The trained CaMelia model can be used for different downstream analyses, including to impute low-coverage methylation profiles for sets of cells or aid the identification of cell types or cell sub-populations.
The overview of CaMelia model
CaMelia does not require installation, just download the necessary files and run from the command line to perform a typical analysis. The code of CaMelia has been tested in Windows and Linux with Python 2.7 (and 3.6).
You will need to download the source code and locate them in the same directory (note: No space characters in the directory path):
a) Feature extraction and datasets formation:
- 6 files in: https://github.com/JxTang-bioinformatics/CaMelia/blob/master/CaMelia%20model/Feature%20extraction
b) Model training and imputing:
Note: The code must be executed in the order described in "Getting started".
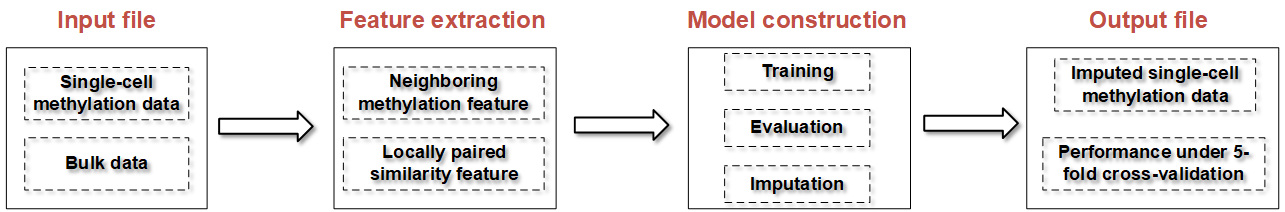
The pipline of the typical CaMelia analysis
1) Store the raw data from cells of the same cell type or bulk data of this cell type into a file (File naming style: "xxx_cell-type.txt", like "GSE65364_mESC.txt") with the following columns:
- Chromosome (with chr)
- Position of the CpG site on the chromosome starting with one
- Binary methylation state of the CpG sites in cell 1 (0=unmethylation, 1=methylated)
- ...
- Binary methylation state of the CpG sites in cell n
- Binary methylation state of the CpG sites in bulk data
The sample input file ("GSE65364_mESC.txt") can be downloaded from: https://github.com/JxTang-bioinformatics/CaMelia/blob/master/example
Example:
chrom location cell1 cell2 ... celln bulk
chr1 136409 1.0 NA ... 0.0 1.0
chr1 136423 NA 1.0 ... NA 0.0
chr1 136425 0.0 NA ... 1.0 1.0
... ...
chrY 28748270 NA 1.0 ... 1.0 1.0
chrY 28748361 0.0 NA ... 0.0 0.0
chrY 28773349 NA NA ... 0.0 0.0
2) Run get_local_Feature_for_train.py and get_neighbor_Feature_for_train.py to extract features for training:
python get_local_Feature_for_train.py Datafilepath(user settings) InputDataName(user settings) LocalRange(user settings: Default 10) CorrelationThreshold(user settings:Default 0.8)
python get_neighbor_Feature_for_train.py Datafilepath(user settings) InputDataName(user settings) LocalRange(user settings:Default 10)
Run get_local_Feature_for_imputation.py and get_neighbor_Feature_for_imputation.py to extract features for imputation:
python get_local_Feature_for_imputation.py Datafilepath(user settings) InputDataName(user settings) LocalRange(user settings:Default 10) CorrelationThreshold(user settings:Default 0.8)
python get_neighbor_Feature_for_imputation.py Datafilepath(user settings) InputDataName(user settings) LocalRange(user settings:Default 10)
3) Run unionfeature_for_train.py and unionfeature_for_imputation.py to generate the datasets for model training and imputing:
python unionfeature_for_train.py Datafilepath(user settings) InputDataName(user settings) LocalRange(user settings:Default 10)
python unionfeature_for_imputation.py Datafilepath(user settings) InputDataName(user settings) LocalRange(user settings:Default 10)
4) Run model_TrainingandImputing.py to train CaMelia, evaluate model performances and impute methylation profiles:
python model_TrainingandImputing.py Datafilepath(user settings) InputDataName(user settings) task_type(user settings:'CPU' or 'GPU')
The main derived output files of CaMelia are:
1) imputation_data: the imputed single-cell methylation data;
2) 5fold_crossvalidation_catboost.csv: the prediction performance of CaMelia models under 5-fold cross-validation;
3) model_save: independent CaMelia models for each cell;
4) model_feature_importance: feature importance Diagram for each independent CaMelia model.
- Jianxiong Tang
- [email protected]