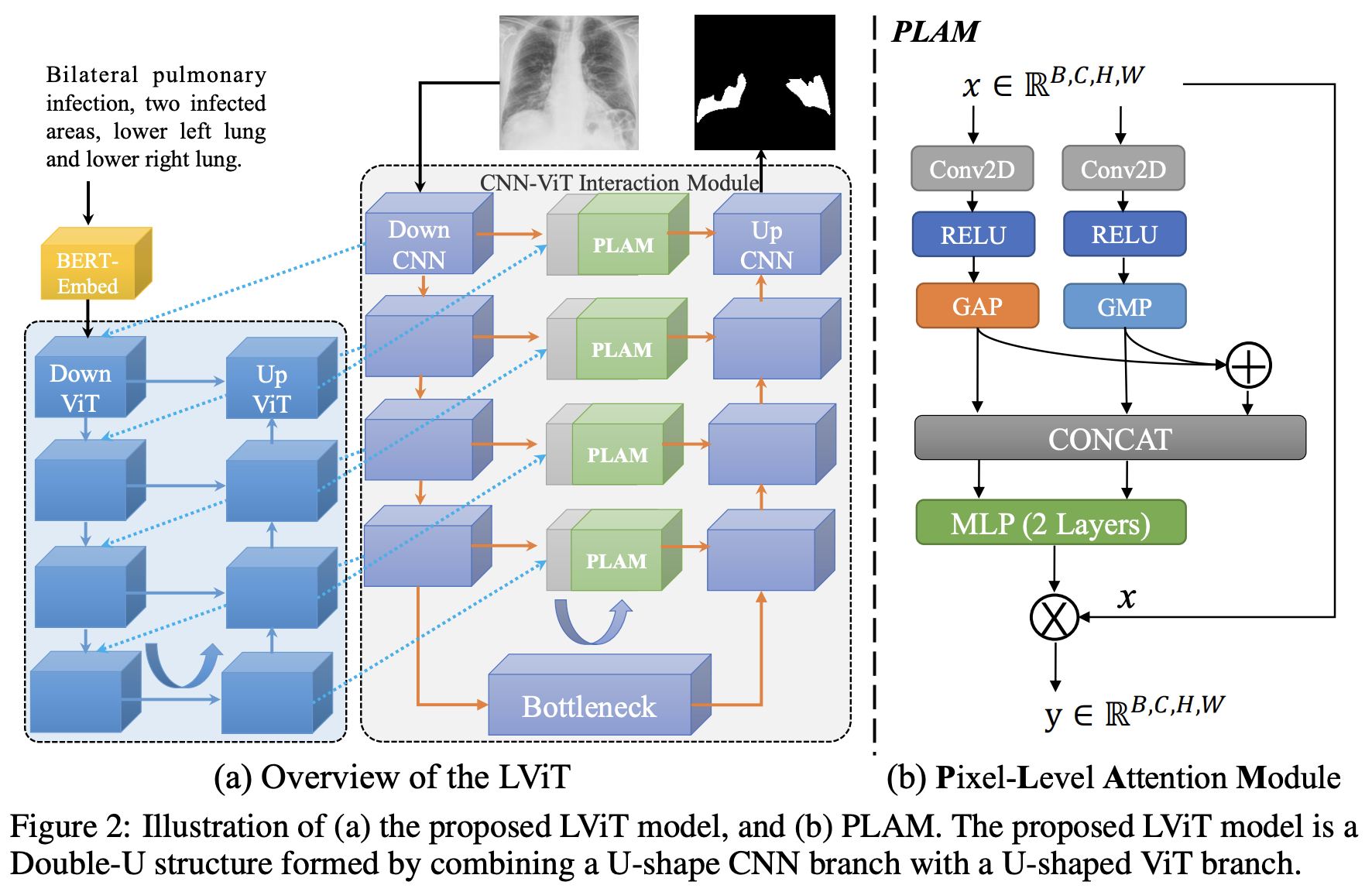
This repo is the official implementation of "LViT: Language meets Vision Transformer in Medical Image Segmentation" Arxiv, ResearchGate, IEEEXplore
Python == 3.7 and install from the requirements.txt using:
pip install -r requirements.txt
Questions about NumPy version conflict. The NumPy version we use is 1.17.5. We can install bert-embedding first, and install NumPy then.
The original data can be downloaded in following links:
-
QaTa-COV19 Dataset - Link (Original)
-
MosMedData+ Dataset - Link (Original) or Kaggle
-
MoNuSeG Dataset (demo dataset) - Link (Original)
-
ESO-CT Dataset [1] [2]
[1] Jin, Dakai, et al. "DeepTarget: Gross tumor and clinical target volume segmentation in esophageal cancer radiotherapy." Medical Image Analysis 68 (2021): 101909.
[2] Ye, Xianghua, et al. "Multi-institutional validation of two-streamed deep learning method for automated delineation of esophageal gross tumor volume using planning CT and FDG-PET/CT." Frontiers in Oncology 11 (2022): 785788.
The text annotation of QaTa-COV19 has been released!
(Note: The text annotation of QaTa-COV19 train and val datasets download link. The partition of train set and val set of QaTa-COV19 dataset download link. The text annotation of QaTa-COV19 test dataset download link.)
(Note: The contrastive label is available in the repo.)
(Note: The text annotation of MosMedData+ train dataset download link. The text annotation of MosMedData+ val dataset download link. The text annotation of MosMedData+ test dataset download link.)
If you use the datasets provided by us, please cite the LViT.
Then prepare the datasets in the following format for easy use of the code:
├── datasets
├── QaTa-Covid19
│ ├── Test_Folder
| | ├── Test_text.xlsx
│ │ ├── img
│ │ └── labelcol
│ ├── Train_Folder
| | ├── Train_text.xlsx
│ │ ├── img
│ │ └── labelcol
│ └── Val_Folder
| ├── Val_text.xlsx
│ ├── img
│ └── labelcol
└── MosMedDataPlus
├── Test_Folder
| ├── Test_text.xlsx
│ ├── img
│ └── labelcol
├── Train_Folder
| ├── Train_text.xlsx
│ ├── img
│ └── labelcol
└── Val_Folder
├── Val_text.xlsx
├── img
└── labelcol
You can replace LVIT with U-Net for pre training and run:
python train_model.py
You can train to get your own model. It should be noted that using the pre-trained model in the step 2.1 will get better performance or you can simply change the model_name from LViT to LViT_pretrain in config.
python train_model.py
First, change the session name in Config.py as the training phase. Then run:
python test_model.py
You can get the Dice and IoU scores and the visualization results.
| Dataset | Model Name | Dice (%) | IoU (%) |
|---|---|---|---|
| QaTa-COV19 | U-Net | 79.02 | 69.46 |
| QaTa-COV19 | LViT-T | 83.66 | 75.11 |
| MosMedData+ | U-Net | 64.60 | 50.73 |
| MosMedData+ | LViT-T | 74.57 | 61.33 |
| MoNuSeg | U-Net | 76.45 | 62.86 |
| MoNuSeg | LViT-T | 80.36 | 67.31 |
| MoNuSeg | LViT-T w/o pretrain | 79.98 | 66.83 |
| Dataset | Model Name | Dice (%) | IoU (%) |
|---|---|---|---|
| BKAI-Poly | LViT-TW | 92.07 | 80.93 |
| ESO-CT | LViT-TW | 68.27 | 57.02 |
In our code, we carefully set the random seed and set cudnn as 'deterministic' mode to eliminate the randomness. However, there still exsist some factors which may cause different training results, e.g., the cuda version, GPU types, the number of GPUs and etc. The GPU used in our experiments is 2-card NVIDIA V100 (32G) and the cuda version is 11.2. And the upsampling operation has big problems with randomness for multi-GPU cases. See https://pytorch.org/docs/stable/notes/randomness.html for more details.
@article{li2023lvit,
title={Lvit: language meets vision transformer in medical image segmentation},
author={Li, Zihan and Li, Yunxiang and Li, Qingde and Wang, Puyang and Guo, Dazhou and Lu, Le and Jin, Dakai and Zhang, You and Hong, Qingqi},
journal={IEEE Transactions on Medical Imaging},
year={2023},
publisher={IEEE}
}