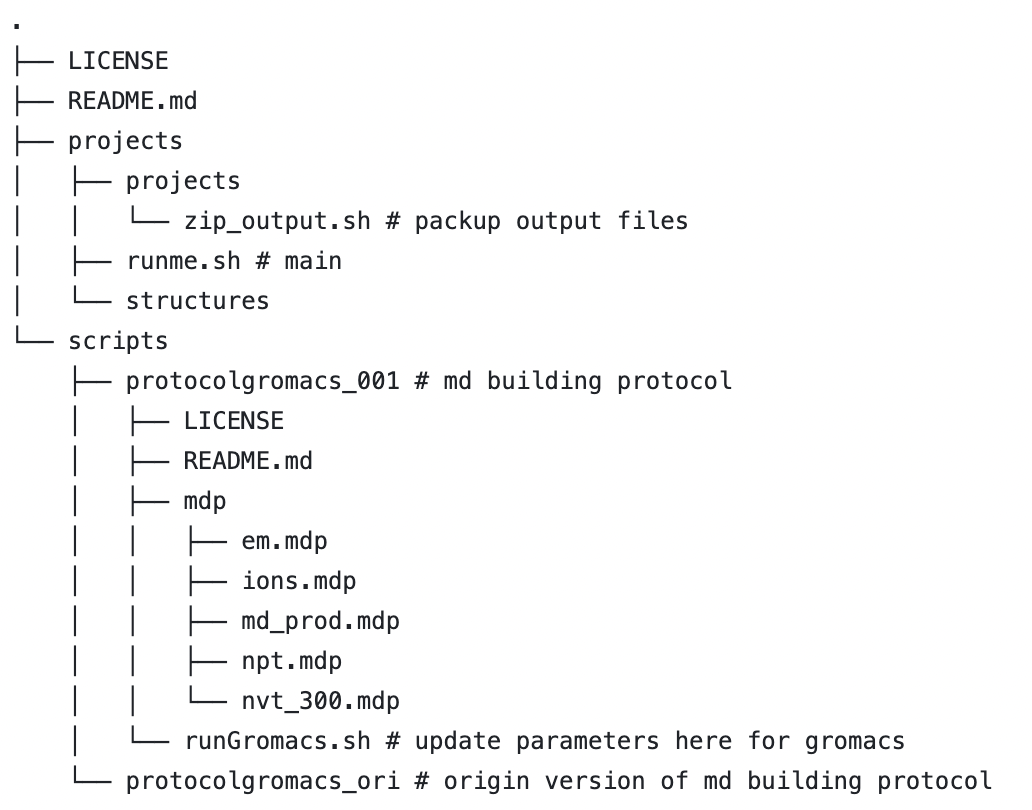
Homemade scripts based on protocolGromacs, for automatic md system building.
Please visit the following links for more information about installation.
The working directory is 'ROOT/projects/'. The script 'runme.sh' will first check PDB files in 'ROOT/projects/structures', then create new directories under 'projects/projects' for each structure, and perform MD system building, minimization, NVT equilibration, NPT equilibration, and so on, as set up in 'ROOT/scripts/protocolgromacs_single_ligand'.
-
Refine PDB files and copy them to 'projects/structures'.
-
Update parameters for GROMACS in 'scripts/protocolgromacs_single_ligand':
- 'ROOT/scripts/protocolgromacs_single_ligand/runGromacs.sh'
- 'ROOT/scripts/protocolgromacs_single_ligand/mdp/*.mdp'
-
Change the directory to 'projects/'.
-
Run the script 'runme_single_protein_single_ligand.sh'.
########################
# for pure protein
########################
# prepare scripts
git clone https://github.com/GiantFurosemide/AutoMdBuilder.git
cp -r ./AutoMdBuilder /path/to/work/
# define ROOT and work directory (not necessary for production)
export MY_ROOT=/path/to/work/AutoMdBuilder
export MY_WORK_DIR=$MY_ROOT/projects
# update mdp files (temperature\timestep\constraint...)
#cd $MY_ROOT/scripts/protocolgromacs_multiple_ligands
#...
# copy prepared structure files to $MY_WORK_DIR/structures
mkdir -p $MY_WORK_DIR/structures
cp -v pure-protein_1.pdb $MY_WORK_DIR/structures/
cp -v pure-protein_2.pdb $MY_WORK_DIR/structures/
cp -v pure-protein_3.pdb $MY_WORK_DIR/structures/
...
# to work directory, run!
cd $MY_WORK_DIR
source runme_single_protein_single_ligand.sh
########################
# for protein + single ligand
########################
# prepare scripts
...
# define ROOT and work directory (not necessary for production)
...
# update mdp files (temperature\timestep\constraint...)
...
# copy prepared structure files to $MY_WORK_DIR/structures
mkdir -p $MY_WORK_DIR/structures
cp -v complex_1.pdb $MY_WORK_DIR/structures/
cp -v complex_2.pdb $MY_WORK_DIR/structures/
cp -v complex_3.pdb $MY_WORK_DIR/structures/
# to work directory, run!
# cause all ligand info defined in runme_single_protein_single_ligand.sh
# complex_1.pdb,complex_2.pdb,complex_3.pdb suposed to contain same ligands
# if complexes consist of different type of ligands,please use 'runme_single_protein_multiple_ligands.sh'
cd $MY_WORK_DIR
source runme_single_protein_single_ligand.sh
Prepare structure files and scripts and copy them to 'ROOT/projects/structures/case1, case2, case3...'. Each sub-directory should contain the following files( for example case1 is an MD system consisting of one protein and two types of ligands, each type with ten ligands):
- complex.pdb (protein-ligand complex pdb file)
- ligand1_GMX.itp, ligand2_GMX.itp ( itp file generated by acpype)
- ligand1_NEW.pdb, ligand2_NEW.pdb (PDB file generated by acpype)
- ligand_info_prepare.py (update ligand info)
- mdbuild_add_multi_ligands.sh(update md system info)
- Update parameters for GROMACS in 'scripts/protocolgromacs_multiple_ligands':
- 'ROOT/scripts/protocolgromacs_multiple_ligands/mdp/*.mdp'
- Change the directory to 'ROOT/projects/'.
- Run the script 'runme_single_protein_multiple_ligands.sh'.
# prepare scripts
git clone https://github.com/GiantFurosemide/AutoMdBuilder.git
cp -r ./AutoMdBuilder /path/to/work/
# define ROOT and work directory (not necessary for production)
export MY_ROOT=/path/to/work/AutoMdBuilder
export MY_WORK_DIR=$MY_ROOT/projects
# update mdp files (temperature\timestep\constraint...)
#cd $MY_ROOT/scripts/protocolgromacs_multiple_ligands
#...
# copy prepared structure files and scripts to 'case1' under $MY_WORK_DIR/structures
mkdir -p $MY_WORK_DIR/structures/case1
cp -v complex.pdb $MY_WORK_DIR/structures/case1
cp -v ligand1_GMX.itp $MY_WORK_DIR/structures/case1
cp -v ligand2_GMX.itp $MY_WORK_DIR/structures/case1
cp -v ligand1_NEW.pdb $MY_WORK_DIR/structures/case1
cp -v ligand2_NEW.pdb $MY_WORK_DIR/structures/case1
cp -v ligand_info_prepare.py $MY_WORK_DIR/structures/case1
cp -v mdbuild_add_multi_ligands.sh $MY_WORK_DIR/structures/case1
# to work directory, run!
cd $MY_WORK_DIR
source runme_single_protein_multiple_ligands.sh
- update supporting for multiple ligands
- update logging