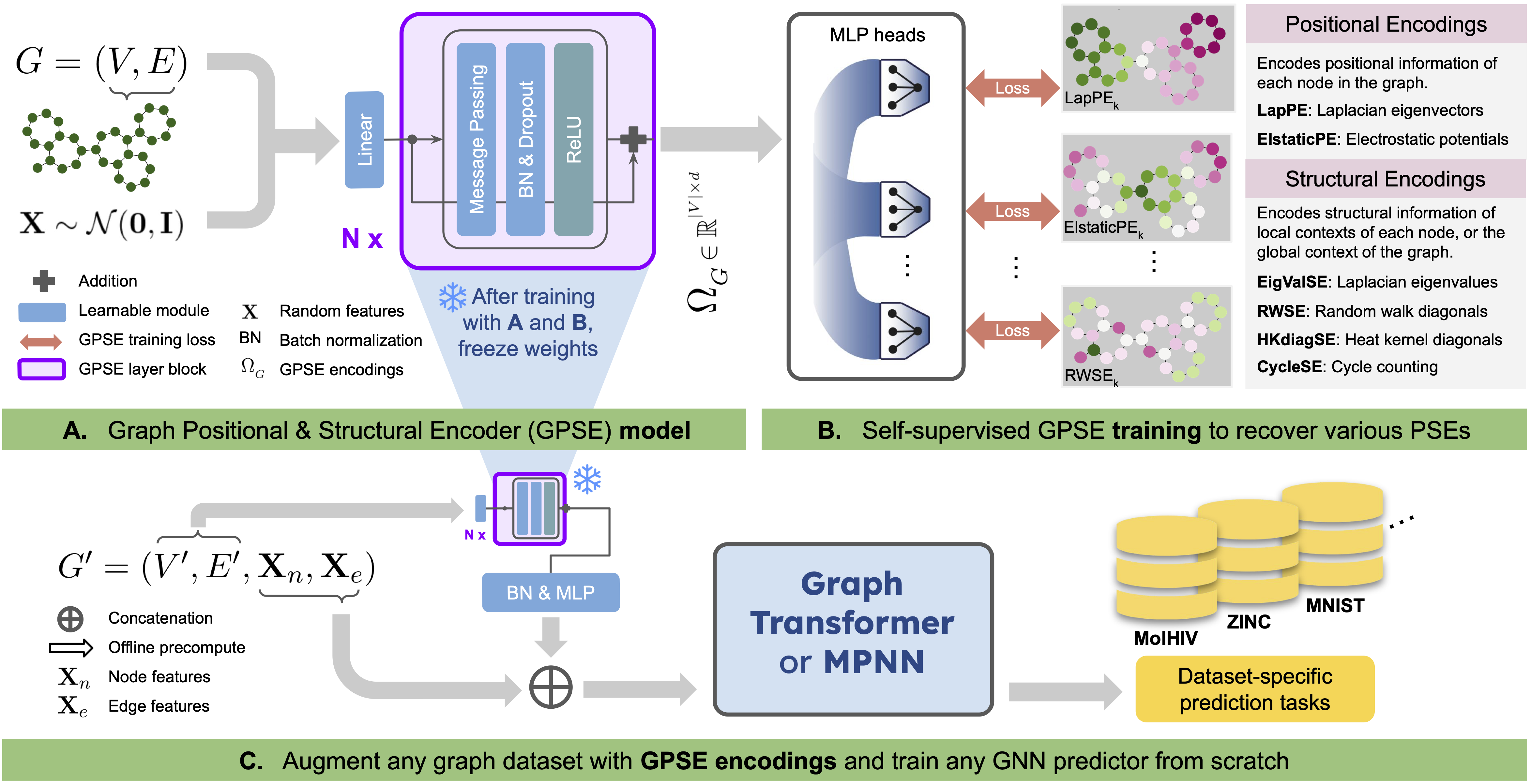
Semih Cantürk*, Renming Liu*, Olivier Lapointe-Gagné, Vincent Létourneau, Guy Wolf, Dominique Beaini, Ladislav Rampášek
Accepted at ICML 2024
This codebase is built on top of GraphGym and GraphGPS. Follow the steps below to set up dependencies, such as PyTorch and PyG:
# Create a conda environment for this project
conda create -n gpse python=3.10 -y && conda activate gpse
# Install main dependencies PyTorch and PyG
conda install pytorch=1.13 torchvision torchaudio pytorch-cuda=11.7 -c pytorch -c nvidia -y
conda install pyg=2.2 -c pyg -c conda-forge -y
pip install pyg-lib -f https://data.pyg.org/whl/torch-1.13.0+cu117.html
# RDKit is required for OGB-LSC PCQM4Mv2 and datasets derived from it.
conda install openbabel fsspec rdkit -c conda-forge -y
# Install the rest of the pinned dependencies
pip install -r requirements.txt
# Clean up cache
conda clean --all -yThe pre-trained GPSE encoder can be downloaded from Zenodo
# Pre-trained on MolPCBA (default)
wget https://zenodo.org/record/8145095/files/gpse_model_molpcba_1.0.pt -O pretrained_models/gpse_molpcba.pt
# Pre-trained on ZINC
wget https://zenodo.org/record/8145095/files/gpse_model_zinc_1.0.pt -O pretrained_models/gpse_zinc.pt
# Pre-trained on PCQM4Mv2
wget https://zenodo.org/record/8145095/files/gpse_model_pcqm4mv2_1.0.pt -O pretrained_models/gpse_pcqm4mv2.pt
# Pre-trained on GEOM
wget https://zenodo.org/record/8145095/files/gpse_model_geom_1.0.pt -O pretrained_models/gpse_geom.pt
# Pre-trained on ChEMBL
wget https://zenodo.org/record/8145095/files/gpse_model_chembl_1.0.pt -O pretrained_models/gpse_chembl.ptYou can also pre-train the GPSE model from scratch using the configs provided, e.g.
python main.py --cfg configs/pretrain/gpse_molpcba.yamlAfter the pre-training is done, you need to manually move the checkpointed model to the pretrained_models/ directory.
The checkpoint can be found under results/gpse_molpcba/<seed>/ckpt/<best_epoch>.pt, where <seed> is the random seed
for this run (0 by default), and <best_epoch> is the best epoch number (you will only have one file, that is the
best epoch).
After you have prepared the pre-trained model gpse_molpcba.pt, you can then run downstream evaluation for models that
uses GPSE encoded features. For example, to run the ZINC benchmark:
python main.py --cfg configs/mol_bench/zinc-GPS+GPSE.yamlYou can also execute batch of runs using the run scripts prepared under run/. For example, to run all molecular
property prediction benchmarks (ZINC-subset, PCQM4Mv2-subset, ogbg-molhiv, and ogbg-molpcba)
sh run/mol_bench.shThis part is for generating the embedding PCA plots in appendix E, Fig. E2.
The plots here show how random initial node features enable breaking symmetries in otherwise 1-WL indistinguishable graphs.
By default, the embeddings are generated by gnn_encoder.py drawn from random normals (see line 32).
To compare with identical input features (e.g. 1), we return a np.ones array of size (n, dim_in), instead of np.random.normal.
Running the code below with and without the changes described above will result in two .pt files of the embeddings.
The code and further instructions to generate the visualizations are found in viz/wl_viz.ipynb.
python viz/wl_test.py --cfg configs/wl_bench/toywl-GPS+GPSE_v9.yaml
- Incompatibility with PyG 2.4.0 due to a minor bug in the GraphGym MLP construction (see #1 and pyg-team/pytorch_geometric#8484).
If you find this work useful, please cite our paper
@misc{liu2023graph,
title={Graph Positional and Structural Encoder},
author={Renming Liu and Semih Cantürk and Olivier Lapointe-Gagné and Vincent Létourneau and Guy Wolf and Dominique Beaini and Ladislav Rampášek},
year={2023},
eprint={2307.07107},
archivePrefix={arXiv},
primaryClass={cs.LG}
}