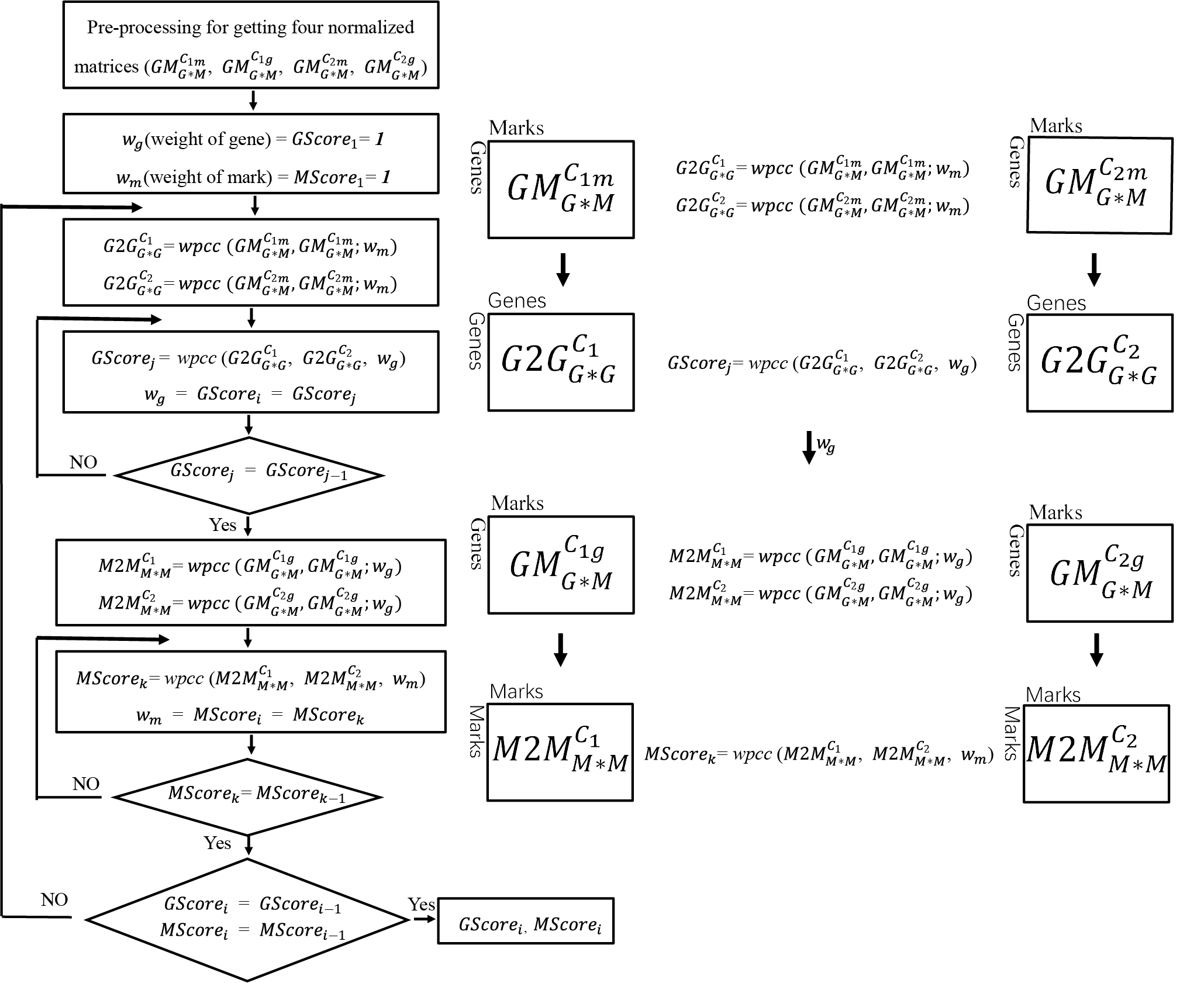
Function: This script is to simultaneously estimate the epigenetic conservation of genes and the conservation of histone modifications between two cell lines or two conditions. It was purely written by R.
parallel;
weights;
scoure ICGEC.R
results<-calculatefun(df1, df2)
ICGEC_score_of_gene <- results[[1]]
ICGEC_score_of_mark <- results[[2]]
The input files (df1 and df2) are two data frames in the form of gene-by-mark, e.g.
H2AK5ac H2BK120ac H2BK5ac H3K18ac H3K23ac H3K27ac H3K27me3 H3K36me3 H3K4ac H3K4me1 H3K4me2 H3K4me3 H3K79me1 H3K9ac H3K9me3 H4K8ac
ENSG00000000003 0.18 0.23 0.23 0.29 0.20 0.77 0.06 1.41 0.17 0.18 1.33 1.98 0.18 1.23 0.23 1.02
ENSG00000000005 0.06 0.50 0.03 0.32 0.29 0.92 0.20 0.11 0.11 0.67 0.74 0.08 0.11 0.22 0.17 0.11
ENSG00000000419 0.07 0.10 0.28 0.17 0.22 0.31 0.08 2.49 0.16 0.28 1.27 2.80 1.33 1.25 0.33 0.59
ENSG00000000457 0.13 0.24 0.30 0.23 0.34 0.55 0.11 2.06 0.20 0.27 0.85 1.42 0.74 0.77 0.35 0.36
ENSG00000000460 0.13 0.27 0.19 0.31 0.34 0.32 0.69 0.92 0.18 0.24 0.40 0.40 0.34 0.54 0.52 0.17
ENSG00000000938 0.94 0.64 0.93 1.56 0.68 0.39 2.96 0.15 0.60 1.30 1.74 0.86 0.28 0.60 0.19 1.60
gene_weight gene_weight.1 gene_weight.2 gene_weight.3 gene_weight.4 gene_weight.5 gene_weight.6 gene_weight.7 gene_weight.8
ENSG00000000003 0.49 0.60 0.61 0.61 0.61 0.60 0.60 0.60 0.60
ENSG00000000005 0.49 0.61 0.63 0.65 0.65 0.66 0.66 0.66 0.66
ENSG00000000419 0.76 0.86 0.87 0.88 0.88 0.88 0.88 0.88 0.88
ENSG00000000457 0.71 0.81 0.82 0.82 0.82 0.82 0.82 0.82 0.82
ENSG00000000460 0.72 0.79 0.77 0.76 0.75 0.75 0.75 0.75 0.75
ENSG00000000938 0.70 0.84 0.85 0.85 0.85 0.85 0.85 0.85 0.85
modi_weight modi_weight.1 modi_weight.2 modi_weight.3 modi_weight.4 modi_weight.5 modi_weight.6 modi_weight.7 modi_weight.8
H2AK5ac 0.67 0.67 0.66 0.66 0.66 0.66 0.66 0.66 0.66
H2BK120ac 0.69 0.76 0.79 0.81 0.82 0.82 0.82 0.83 0.83
H2BK5ac 0.49 0.53 0.57 0.63 0.66 0.67 0.67 0.68 0.68
H3K18ac 0.80 0.84 0.85 0.85 0.85 0.85 0.85 0.85 0.85
H3K23ac 0.54 0.52 0.52 0.52 0.53 0.53 0.54 0.54 0.54
H3K27ac 0.62 0.60 0.59 0.59 0.59 0.59 0.60 0.60 0.60
H3K27me3 0.79 0.83 0.84 0.85 0.85 0.85 0.85 0.85 0.85
H3K36me3 0.76 0.81 0.82 0.83 0.83 0.83 0.84 0.84 0.84
H3K4ac 0.67 0.72 0.74 0.76 0.77 0.77 0.77 0.77 0.77
H3K4me1 0.45 0.39 0.35 0.32 0.30 0.28 0.26 0.25 0.23
H3K4me2 0.85 0.87 0.87 0.87 0.87 0.87 0.87 0.87 0.87
H3K4me3 0.80 0.84 0.85 0.86 0.86 0.86 0.86 0.86 0.86
H3K79me1 0.86 0.87 0.87 0.87 0.87 0.87 0.87 0.87 0.87
H3K9ac 0.82 0.83 0.84 0.84 0.83 0.83 0.83 0.83 0.83
H3K9me3 0.72 0.76 0.76 0.75 0.74 0.74 0.74 0.73 0.73
H4K8ac 0.31 0.17 0.04 -0.11 -0.19 -0.20 -0.21 -0.21 -0.21
#Note: Columns represent the results from the iterative process of ICGEC. The last column is the final converged scores (ICGEC score of genes or marks).