This repository is the PyTorch implementation of MolR (paper):
Chemical-Reaction-Aware Molecule Representation Learning
Hongwei Wang, Weijiang Li, Xiaomeng Jin, Kyunghyun Cho, Heng Ji, Jiawei Han, Martin Burke
The 10th International Conference on Learning Representations (ICLR 2022)
MolR uses graph neural networks (GNNs) as the molecule encoder, and preserves the equivalence of molecules w.r.t. chemical reactions in the embedding space.
Specifically, MolR forces the sum of the reactant embeddings and the sum of the product embeddings to be equal for each chemical reaction, which is shown to keep the embedding space well-organized and improve the generalization ability of the model.
MolR achieves substantial gains over state-of-the-art baselines.
Below is the result of Hit@1 on USPTO-479k and real reaction dataset for the task of chemical reaction prediction:
| Dataset | USPTO-479k | real reaction |
|---|---|---|
| Mol2vec | 0.614 | 0.313 |
| MolBERT | 0.623 | 0.313 |
| MolR-TAG | 0.882 | 0.625 |
Below is the result of AUC on BBBP, HIV, and BACE datasets for the task of molecule property prediction:
| Dataset | BBBP | HIV | BACE |
|---|---|---|---|
| Mol2vec | 0.872 | 0.769 | 0.862 |
| MolBERT | 0.762 | 0.783 | 0.866 |
| MolR-GCN | 0.890 | 0.802 | 0.882 |
Below is the result of RMSE on QM9 dataset for the task of graph-edit-distance prediction:
| Dataset | QM9 |
|---|---|
| Mol2vec | 0.995 |
| MolBERT | 0.937 |
| MolR-SAGE | 0.817 |
Below are the visualized reactions of alcohol oxidation and aldehyde oxidation using PCA:

Below is the visualized molecule embedding space on BBBP dataset using t-SNE:
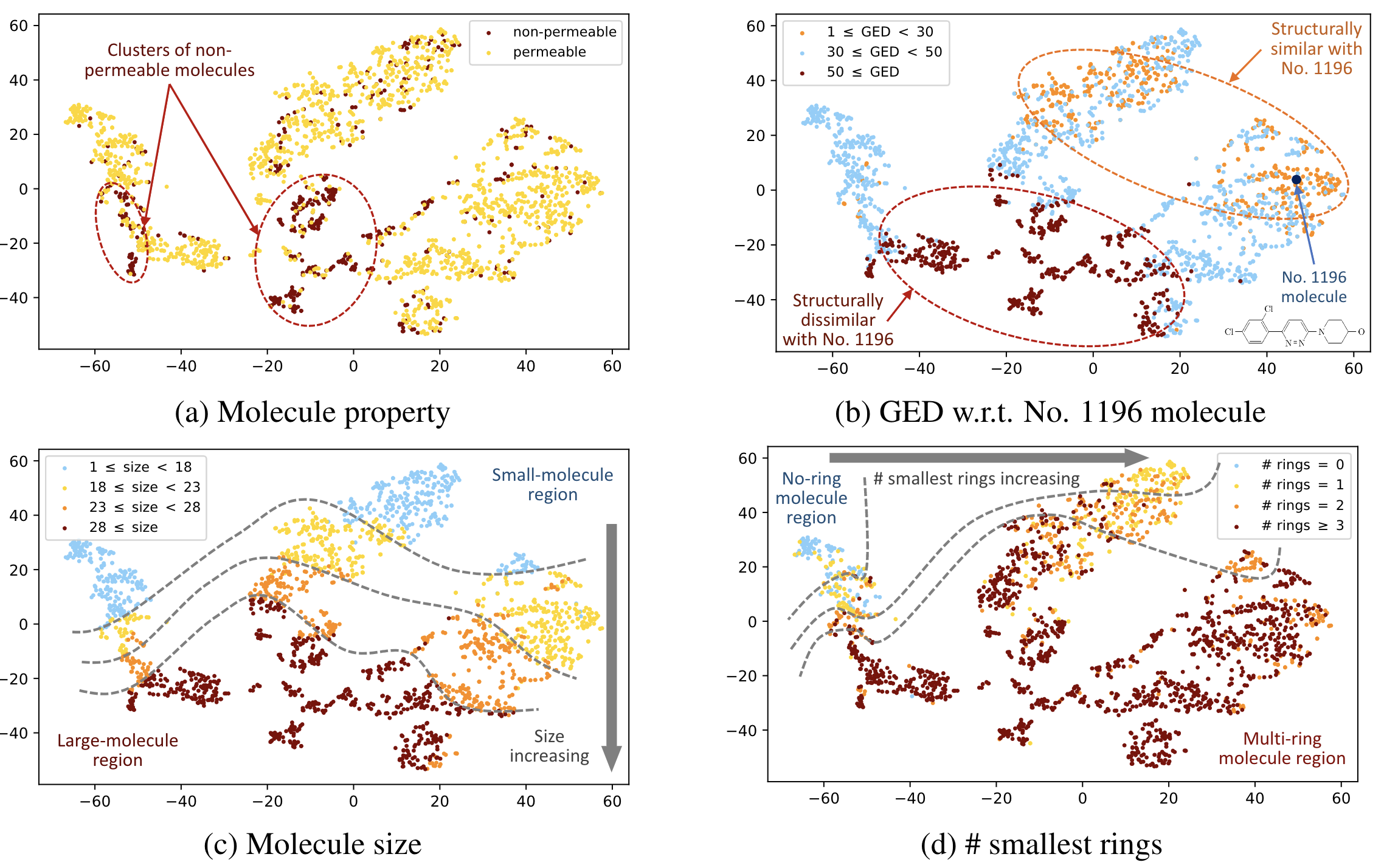
For more results, please refer to our paper.
data/USPTO-479k/for chemical reaction predictionUSPTO-479k.zip: zipped file containingtrain.csv,valid.csv, andtest.csv. Please unzip this file and put the three csv files under this directory. The cached files of USPTO-479k in DGL format is too large to be uploaded to GitHub. If you want to save the time of pre-processing this dataset, please download them here and put the unzipped/cachedirectory under this directory.
real_reaction_test/for chemical reaction predictionreal_reaction_test.csv: SMILES of reactants and multiple choices of 16 real questions
BBBP/for molecule property prediction and visualizationBBBP.csv: the original dataset fileBBBP.bin: the cached dataset in DGL formatged_wrt_1196.pkl: the cached graph-edit-distance of all molecules w.r.t. No. 1196 moleculesssr.pkl: the cached numbers of smallest rings for all molecules
HIV/for molecule property predictionHIV.csv: the original dataset file. Note that theHIV.binis too large to be uploaded to GitHub. If you want to save the time of pre-processing this dataset, please download it here and put the unzippedHIV.binunder this directory.
BACE/for molecule property predictionBACE.csv: the original datasetBACE.bin: the cached dataset in DGL format
Tox21/for molecule property predictionTox21.csv: the original datasetTox21.bin: the cached dataset in DGL format
ClinTox/for molecule property predictionClinTox.csv: the original datasetClinTox.binthe cached dataset in DGL format
QM9/for graph-edit-distance predictionQM9.csv: the original datasetpairwise_ged.csv: the randomly selected 10,000 molecule pairs from the first 1,000 molecules in QM9.csvged0.binandged1.bin: cached molecule pairs in DGL format
src/ged_pred/: code for graph-edit-distance prediction taskproperty_pred/: code for molecule property prediction taskreal_reaction_test/: code for real reaction test taskvisualization/: code for embedding visualization taskdata_processing.py: processing USPTO-479k datasetfeaturizer.py: an easy-to-use API for converting a molecule SMILES to embeddingmain.py: main functionmodel.py: implementation of GNNstrain.py: training procedure on USPTO-479k dataset
saved/(pretrained models with the name format ofgnn_dim)gat_1024/gcn_1024/sage_1024/tag_1024/
- If you would like to train the model from scratch or evaluate the pretrained model on downstream tasks of molecule property prediction, GED prediction, or visualization, just uncomment the corresponding part of code in
main.pyand runThe default task is set as$ python main.py--task=pretrain. Hyper-parameter settings for other tasks are provided inmain.py, which should be easy to understand.
Note: If you spot CUDA_OUT_OF_MEMORY error under the default hyperparameter setting, please consider decreasing the batch size first (e.g., to 2048). This will decrease the performance but not too much. - If you would like to use the pretrained model to process your own molecules and obtain their embeddings, please run
Please see
$ python featurizer.pyexample_usage()function infeaturizer.pyfor details, which should be easy to understand.
The code has been tested running under Python 3.7 and CUDA 11.0, with the following packages installed (along with their dependencies):
- torch == 1.8.1
- dgl-cu110 == 0.6.1 (can also use dgl == 0.6.1 if run on CPU only)
- pysmiles == 1.0.1
- scikit-learn==0.24.2
- networkx==2.5.1
- matplotlib==3.4.2 (for
--task=visualization) - openbabel==3.1.1 (for
--task=visualizationand--subtask=ring) - scipy==1.7.0