PEANuT is a Cytoscape 2.8.3 plugin for pathway enrichment analysis.
PEANuT utilizes data from the following databases:
The version for Cytoscape 3.x is available via https://sourceforge.net/projects/peanutv3/
PEANuT is designed to work in concert with the VIPER and COMFI plugins.
If you use PEANuT please cite:
- Internet connection
- Java 1.5+
- Cytoscape 2.8
In general the installation procedure comprises 2 simple, quick steps:
- Download and install PEANuT in Cytoscape
- Set up PEANuT through the “Download/update dependencies” submenu
There are three easy, different ways to install PEANuT in Cytoscape:
- Search for the PEANuT plugin under the Cytoscape menu “plugins” “manage plugins” and click on install.
- Download the PEANuT.jar file from the project page and click on “Plugins” “Install plugin from file” and select the downloaded .jar file.
- Download the PEANuT.jar file from the project page and place it in the “plugins” folder in your Cytoscape installation directory.
 If the plugin is correctly installed you should now be able to see a new “PEANuT” entry in the “plugins menu”. If that is not the case try restarting Cytoscape.
If the plugin is correctly installed you should now be able to see a new “PEANuT” entry in the “plugins menu”. If that is not the case try restarting Cytoscape.
In order to work properly PEANuT requires several files from the ConsensusPathDB and WikiPathways databases. To download these files go to the “Download/update dependencies” submenu in the “PEANuT” entry.
You should see a dialogue like below pop up. Just click on the symbol in the red circle and download all files. This should only take a few seconds. When the download is completed you will be notified of a successful download and the status icon will switch from the red cross to a green tick. This means that PEANuT is now ready to use!
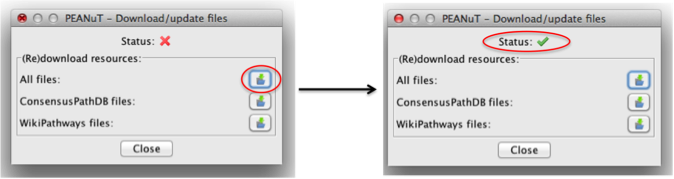
If you have no access to the internet or can't download the files due to server issues from the databases you can also download a zipped archive here. After downloading simply unzip the contents into you Cytoscape plugin folder. However the zip file might contain slightly older file versions.
- clone the git repository to your computer
- open the build.properties
- set the following two variables: ..* classpath.local.location=TheFolderThatContainsThisSourceCode/lib ..* cytoscape.plugins.path=YourCytoscapeFolder/plugins/
- make sure you have ant installed
- run
ant build.xmlto compile the plugin
The PEANuT menu comprises the submenus:
The menu entry “Find pathways” allows the user to annotate the nodes of a Cytoscape session with information from the three different pathway databases ConsensusPathDB, Pathway Commons and WikiPathways.
After clicking on the “Find pathways” submenu a dialogue as displayed in the figure below will pop up. There you have the possibility so select the database of your choice via the 3 tabs displayed on top of the dialogue. After selecting a database you will be provided with several other options that need to be set according to your data and preferences.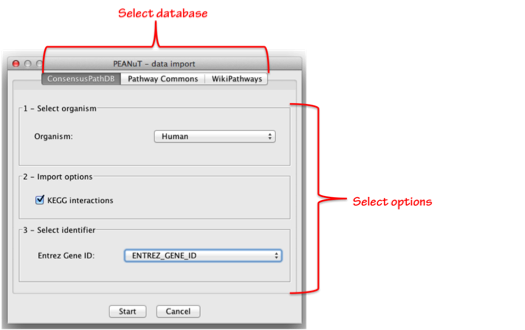
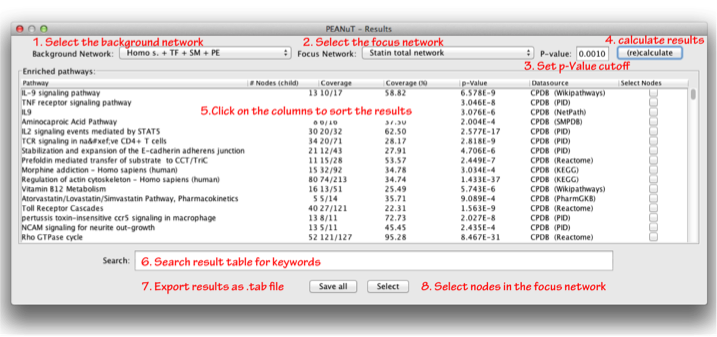
The menu entry “Show results” allows the user to browse the results of the pathway annotation procedure. When clicking on the “Show results” submenu a dialogue as displayed in the figure below will pop up. There you have the possibility so select to e.g. select a background and focus network, a cutoff p-Value for enrichment testing or to export the results as a tab separated file.
- Select the background network: Select the network which will be the background for the statistical testing, e.g. Yeast or Human interactome. The default is the largest network present in your session.
- Select the focus network: Select the network for which you want to know if there are any pathways enriched.
- Set p-Value cutoff: You can enter p-Value cutoff. Only pathways with a smaller p-Value than the cutoff value will be displayed. The default is 0.001. Allowed values range from 1 - 0.
- (Re)calculate results: When clicking the calculate results button the application will calculate which pathways are enriched (based on options 1-3).
- Click on the columns to sort the results: You can click on the header of each column to sort it either alphabetically or according to its numeric value. The columns display the names of the pathway, the number of nodes in the fous network associated with this pathway, the coverage of the pathway (as fraction and percent), the respective p-Value, the datasource of the pathway and an option whether you later want to select the nodes in the focus network or not (see also point 8). Note that the number of nodes and the coverage may differ. This is due to the fact that some networks may contain additional nodes for super pathways, parts of super pathways, catalytic reactions or nodes for different isoforms that are mapped to the same Entrez gene IDs.
- Search result table for keywords: If you are looking for specific pathways you can type keywords in this text field and search the result table (case insensitive).
- Export results as .tab file: Clicking this button will open a file chooser that will allow you to save the results displayed in the table as a tab separated file that can easily be imported in to Excel, OpenOffice etc.
- Select nodes in the focus network: Clicking this button will select the nodes of the ticked pathways (Column select nodes) in the focus network.
If you want to get some hand on experience with PEANuT you can download a test network comprised out of the yeast proteome here and have a look at our walkthrough example below.
Before we can dive into the example you need to:
- Install Cytoscape and PEANuT (see our installation guide)
- Download our example network
After you have started Cytoscape open the example session file via "File" --> "Open" and the select the yestTestNetwork.cys you have just downloaded.
Using the plugin consist of two simple steps:
- Annotate the networks with pathway data
- Calculate and review the results
If you want a detailed overview on how to use the different submenus see here.
To annotate the network with pathway data go to the menu "Plugins"-->"PEANuT"-->"Find pathways". This only needs to be done once for each session file. For this example e.g. select ConsensusPathDB.
- Set the organism to Yeast
- unselect import KEGG interactions
- select "entrezgene" in the combo box
- Click start
To review the results of possible significant enriched pathways go to the menu "Plugins"-->"PEANuT"-->"Show pathway statistics".
- Select "Binary-GS.txt" as background network
- Select "Binary-GS.txt--child" as focus network
- Change the p-Value to you are prefrences or leave it at 0.001
- Click (re)calculate
- If you chose p=0.001 you will see a table comprising of only one entry.
| Pathway | # Nodes (focus) | Coverage | Coverage (%) | p-Value | Datasource |
|---|---|---|---|---|---|
| Trehalose Biosynthesis | 4 | 4/4 | 100.00 | 2.066E-4 | CPDB (Wikipathways) |
You can click on "Save all", save the result as a tab separated file and import it into Excel. This is done in Excel via "File"--> "open", setting the file filter to "enable all documents" and then simply selecting and opening it. Afterwards Excel will ask you in a series of dialogues which kind of delimiter this file uses. Just select tab and "Finish".
If you want to play a bit around with the test network, just create other smaller or larger subnetworks and test for enrichment or annotate the networks with pathway data from other databases (e.g. Pathway Commons or WikiPathways).
You can also ask questions in our google group.

