Epitopes are surface-exposed regions of a pathogenic molecule or antigen, which are targeted by the adaptive immune system by e.g. B-cell receptors. Binding is largely determined by the surface features of the target molecule. AlphaFold2 is a deep-learning protein folding model achieving near experimental quality prediction for many proteins. Furthermore, graph-based neural networks such as ProteinSolver allow structural representation of proteins suitable for tasks such as epitope prediction. We investigate improved epitope prediction using Alphafold2 modelled structures over sequence-only models.
02456 DEEP LEARNING, DTU COMPUTE, FALL 2021
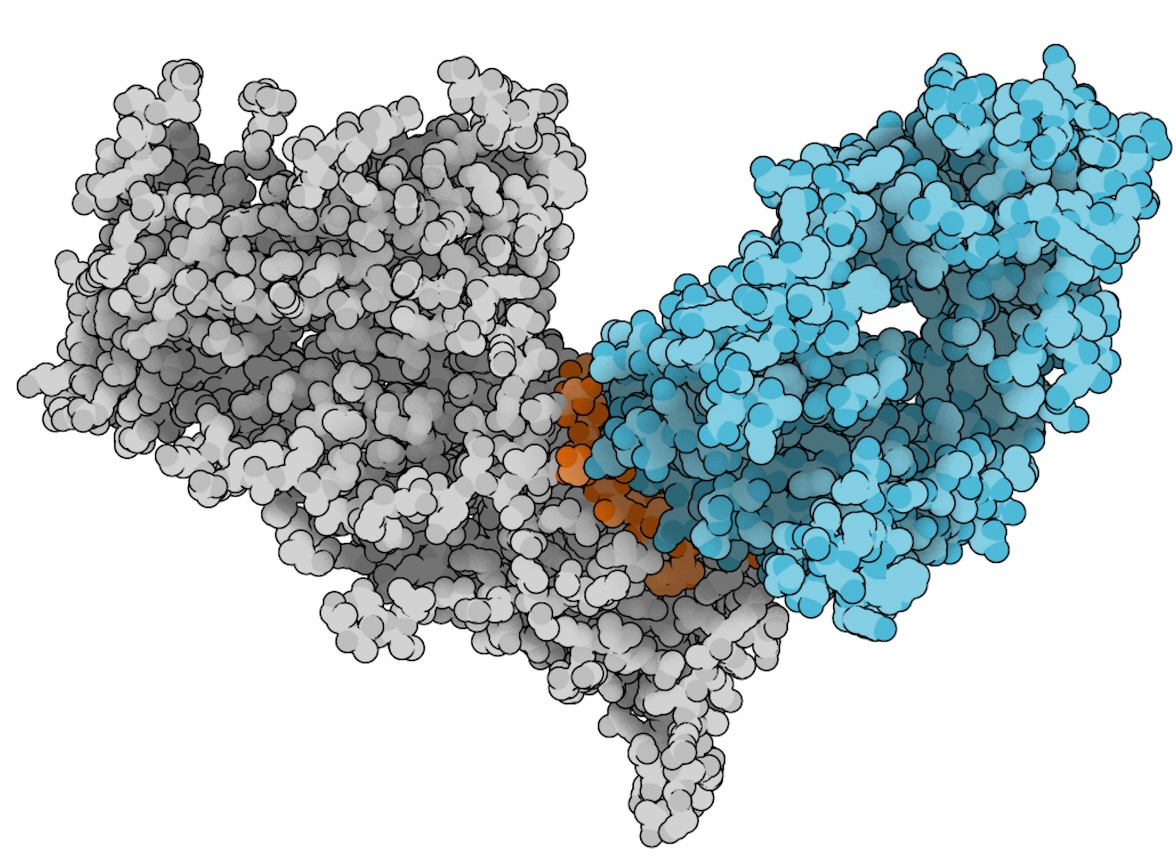
Figure 1: Neutralizing antibody (cyan) bound to the epitope region (orange) of an HIV glycoprotein antigen (grey).
The main notebook of this project, Epitope_Prediction.ipynb, contains demonstrations of our three models (FFNN, RNN & GNN) and two reference models (Discotope2 [1] and BepiPred3 [2]) and a comparison of their performance. We allow use of both solved structure embeddings and AlphaFold2 [3] structure embeddings. Both of these embeddings are made with ProteinSolver [4].
Running this notebook requires specific versions of PyTorch and associated packages to be compatible with ProteinSolver. Using anaconda, the following commands should suffice to setup the correct environment:
conda env create -f PS_gpu.yaml
conda activate PS_gpu
conda install nb_conda
Note that installing nb_conda makes it possible to use the conda environment in jupyter
[1] Kringelum, J. V., Lundegaard, C., Lund, O., & Nielsen, M. (2012). Reliable B Cell Epitope Predictions: Impacts of Method Development and Improved Benchmarking. PLoSComputationalBiology, 8(12). https://doi.org/10.1371/journal.pcbi.1002829
[2] Jespersen, M. C., Peters, B., Nielsen, M., & Marcatili, P. (2017). BepiPred-2.0: Improving sequence-based B-cell epitope prediction using conformational epitopes. NucleicAcidsResearch, 45(W1), W24–W29. https://doi.org/10.1093/nar/gkx346
[3] Jumper, J et al (2021). Highly accurate protein structure prediction with AlphaFold. Nature, May, 1–12. https://doi.org/10.1038/s41586-021-03819-2
[4] Strokach, A., Becerra, D., Corbi-Verge, C., Perez-Riba, A., & Kim, P. M. (2020). Fast and Flexible Protein Design Using Deep Graph Neural Networks. Cell Systems, 11(4), 402-411.e4. https://doi.org/10.1016/j.cels.2020.08.016