Code to support paper in https://www.cell.com/patterns/
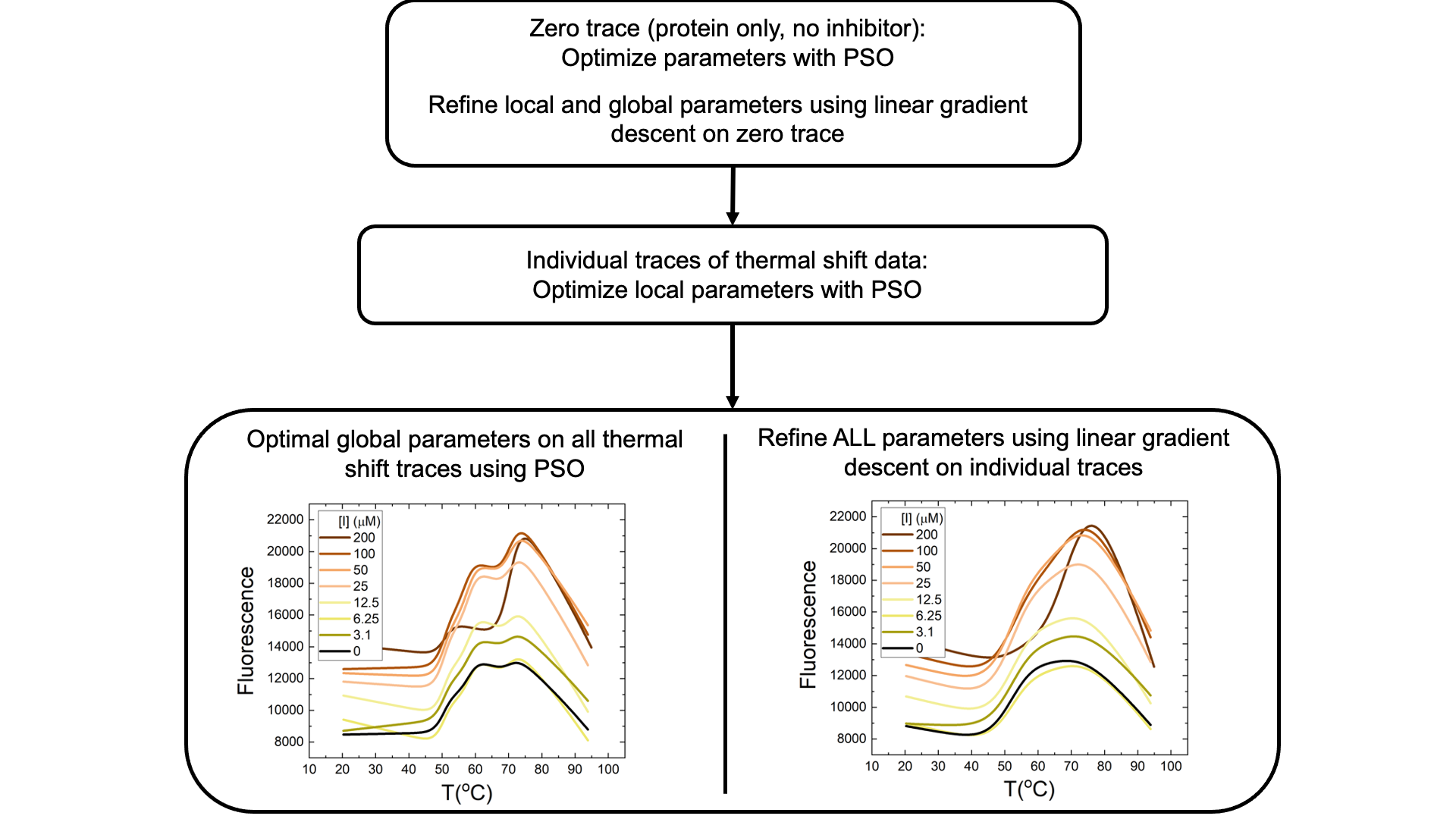
Application of PSO (Particle Swarm Optimization) to understand the allosteric mechanism of action of inhibition of the enzyme HSD17ß13
- Python 3.9
To perform a single run, install the requirements and run the main run.py python file.
pip install -r requirements.txt
python run.py
- Fluorescence data for a “zero trace” (i.e., protein alone, no inhibitor) is analysed and parameters are optimized using PSO.
- Linear gradient descent is performed on the “zero trace” following PSO to refine fitting parameters
- PSO is used on the individual traces (i.e., each inhibitor concentration) of thermal shift data to derive local parameters
- Then, optimal global parameters for all traces are determined using PSO and refined using linear gradient descent
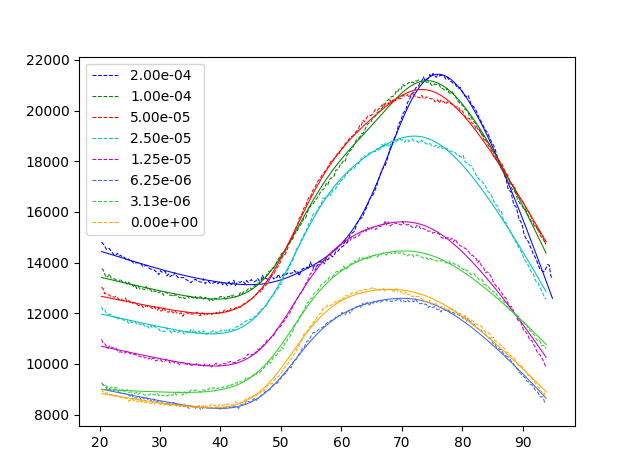
You will see the pyswarms output describing the progress of each PSO step. At the end, 2 plots should be produced of the final 2 fitting steps, the latter looking like the plot below:
Note: A perfect fit is not expected to occur every time. For this dataset, it should reach the correct final solution 15% of the time. See below.
- Multiple runs: If you would like to see the performance of the model over a set of runs, increase
NUMBER_OF_RUNSinrun.pyto e.g. 100. - Saving images/files: If you would like the results of the fits exported to csvs, and the files and plots generated saved to disk, create the export folders
CSV ExportsandPlotsin the home directory and setSAVE_TO_DISKinrun.pytoTrue