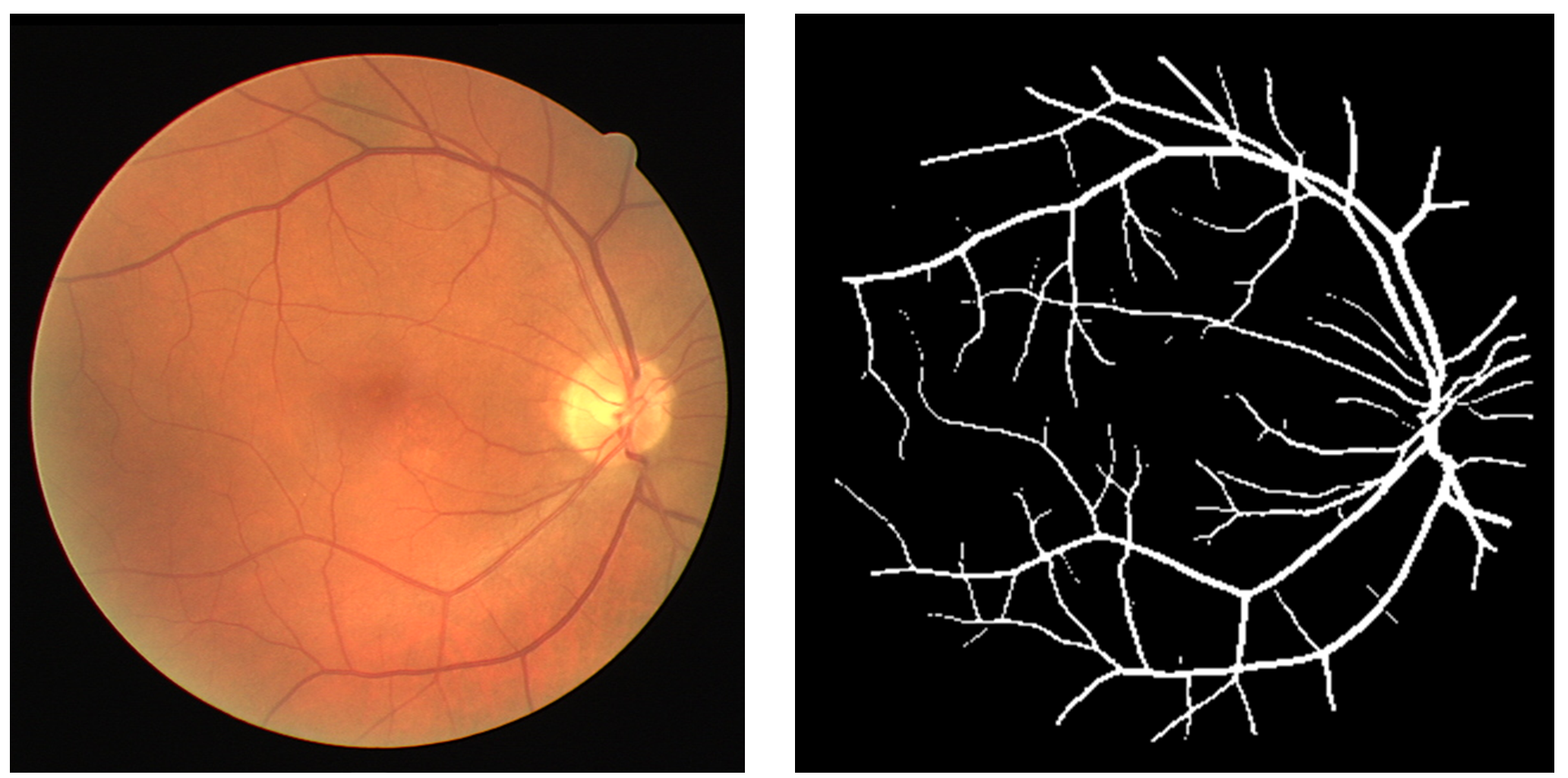
This provides a software package to automatically segment vasculature from Retinal Fundus photographs. The model uses an ensemble of 10 U-Nets trained on datasets from DRIVE, CHASEdb, and STARE.
This inference code was tested on Ubuntu 18.04.3 LTS, conda version 4.8.0, python 3.7.7, fastai 1.0.61, cuda 10.2, pytorch 1.5.1 and cadene pretrained models 0.7.4. A full list of dependencies is listed in environment.yml.
Inference can be run on the GPU or CPU, and should work with ~4GB of GPU or CPU RAM. For GPU inference, a CUDA 10 capable GPU is required.
For the model weights to download, Github's large file service must be downloaded and installed: https://git-lfs.github.com/
This example is best run in a conda environment:
git lfs clone https://github.com/vineet1992/Retina-Seg/
cd location_of_repo
conda env create -n Retina_Seg -f environment.yml
conda activate Retina_Seg
python Code/seg_ensemble.py test_images/ test_output/ ../model/UNet_Ensemble 10Dummy image files are provided in test_images/;. Weights for the segmentation model are in model/UNet_Ensemble_[0-9].pth.
Output will be written to test_output/.
I thank the creators of the DRIVE, CHASEdb, STARE databases along with the UK Biobank for access to their datasets.