Supplementary code and data of the paper GCN2defect: Graph Convolutional Networks for SMOTETomek-based Software Defect Prediction.
@INPROCEEDINGS{9700305, author={Zeng, Cheng and Zhou, Chun Ying and Lv, Sheng Kai and He, Peng and Huang, Jie}, booktitle={2021 IEEE 32nd International Symposium on Software Reliability Engineering (ISSRE)}, title={GCN2defect : Graph Convolutional Networks for SMOTETomek-based Software Defect Prediction}, year={2021}, volume={}, number={}, pages={69-79}, doi={10.1109/ISSRE52982.2021.00020}}
In each subdirectory, we have already included the corresponding Class Dependency Network (CDN) (edges.txt). If you want to generate your own CDN, you can use the Dependencyfinder API.
Before training GCN, we have to provide the attributes of the CDN nodes. Thus, three types of node metrics are introuduced as node attributes:
1) Traditional Static Code Metric: 20 manually designed metrics (Process-Binary.csv).
2) Complex Network Metric: widely used in social network and included 17 metrics (Process-Metric.csv).
3) Network Embedding Metric: use the ProNE implementation to generate the network embedding file (Process-Vector.csv).
Run generateGCNemb.py to generate GCN embeddings.
We use the the stellargraph to generate the GCN embeddings. The GCN demo shows in https://stellargraph.readthedocs.io/en/stable/demos/node-classification/gcn-node-classification.html.
If you want to change to your own dataset, you need the following steps:
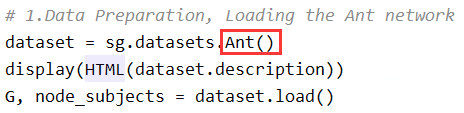
1) Replace the name in the red box in the following figure with the name of your dataset.
2) Place the mouse over the dataset, then press Ctrl, and click to enter init.py.
Add the name of your dataset in init.py.
3) Place the mouse over the dataset name (except for the dataset name you just created), then press Ctrl, and click to enter datasets.py.
4) Create your own class in datasets.py. For example, the following code is to create Ant dataset:
class Ant(
DatasetLoader,
name="Ant",
directory_name="Ant",
url="",
url_archive_format="",
expected_files=[],
description="",
source="",
):
_NUM_FEATURES = 20
def load(
self,
directed=False,
largest_connected_component_only=False,
subject_as_feature=False,
edge_weights=None,
str_node_ids=False,
):
nodes_dtype = str if str_node_ids else int
return _load_defect_data(
self,
directed,
largest_connected_component_only,
subject_as_feature,
edge_weights,
nodes_dtype,
)def _load_defect_data(
dataset,
directed,
largest_connected_component_only,
subject_as_feature,
edge_weights,
nodes_dtype,
):
assert isinstance(dataset, (Ant))
if nodes_dtype is None:
nodes_dtype = dataset._NODES_DTYPE
node_data = pd.read_csv("E:\\gcn2defect\\data\\" + dataset.name + "\\Process-Binary.csv")
edgelist = pd.read_csv(
"E:\\gcn2defect\\data\\" + dataset.name+ "\\edges.txt", sep="\t", header=None, names=["target", "source"], dtype=nodes_dtype
)
node_data.apply(pd.to_numeric, errors='ignore')
# 0 to buggy, 1 to clean
subjects_num = node_data['bug']
label_list = subjects_num.to_list()
labels = []
for i in range(len(label_list)):
if label_list[i] == 1:
labels.append('buggy')
else:
labels.append('clean')
subjects = pd.Series(labels, dtype='str')
cls = StellarDiGraph if directed else StellarGraph
features = node_data.iloc[:, 3:-1]
feature_names = node_data.iloc[:, 2]
minMax = preprocessing.MinMaxScaler()
features_std = minMax.fit_transform(features)
graph = cls({"class": features_std}, {"to": edgelist})
if edge_weights is not None:
# A weighted graph means computing a second StellarGraph after using the unweighted one to
# compute the weights.
edgelist["weight"] = edge_weights(graph, subjects, edgelist)
graph = cls({"class": node_data[feature_names]}, {"to": edgelist})
if largest_connected_component_only:
cc_ids = next(graph.connected_components())
return graph.subgraph(cc_ids), subjects[cc_ids]
return graph, subjectsAfter generating the gcn_emb.emd file, we can run the experiment by executing pipeline.py.
python==3.7
stellargraph==1.2.1
tensorflow-gpu==2.0.1
scikit-learn==1.0.2
networkx==2.6.3