This repository contains the code of my Senior Thesis in which I explore how Gated Graph Neural Networks can be used to predict class-level imports in Java code. More specifically, I attempt to predict which classes (or, more properly, compilation units) defined in a certain project currently under development might be imported to a class newly defined in the same project given the new class's name and possibly initial import statements in that class.
Because the goal is to predict imports occurring within a possibly unfinished project, one cannot gather enough import co-occurance statistics to efficiently predict class-level imports. Instead, one must rely exclusively on information that is locally available. To approach this problem, I propose to model relationships between compilation units in a project with a graph. A node in such a graph corresponds to a particular compilation unit. An edge between two nodes marks some relationship between the two corresponding compilation units.
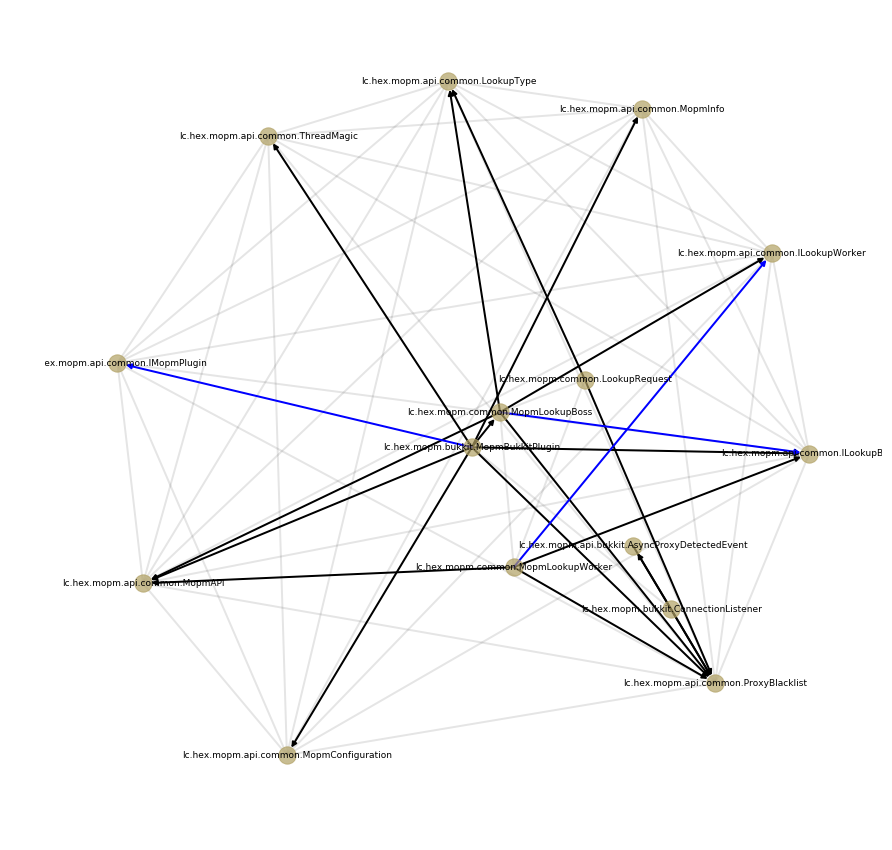
Below is an example of such a graph build for one of the repositories in my dataset. Grey undirected edges connect compilation units defined within the same package. Black directed units correspond to import statements. Blue edges mark class inheritance or interface implementation:
The code in this repository allows building such graphs for arbitrary GitHub projects and running GGNNs on these graphs to learn to predict future imports in a way similar to how Allamanis et al. solve the variable misuse task (they work on the level of a small piece of code and model relationships between variables, while I model relationships between files\compilation units in the context of a repository). The initial comparison between the baseline results I was able to achieve by other means (see notebooks/Baselines.ipynb) suggest that GGNNs are well suited for this task. For more details on the way I use GGNNs, see README.md in the python directory.
The data for this project can be obtained by running a set of SQL queries on publicly available BigQuery Dataset. The README.md file in the SQL directory contains detailed information about replicating this step and the criteria used to select the data.
The raw data can be downloaded from BigQuery via GCS as a set of .json files each of which will contain millions of lines of Java code. This code can be parsed by running Parser.java (located in java/src) passing the name of one .json file as the first argument and the name of the output file as a second argument. You will need JavaParser to run this code.
Next, the data goes through a series of preprocessing steps. To replicate them,
run Filtering.ipynb and ConvertingToGraphs.ipynb notebooks. You might need the
following python libraries to run this code: pytorch, numpy, tqdm, joblib, networkx, sklearn, matplotlib, pandas
More information on data preprocessing and baselines used for import prediction can be found in the README.md file in the notebooks directory.
The detailed desription of the pytorch implementation of GGNNs used for this project can be found in the README.md file in the python directory. The same directory contains the python modules that define the network structure, the loss function, and the way the data loader works. wrapper.py in the python directory contains an example of how the network could be run.