To run Msolve and korali integration example use the docker image,
$ docker build github.com/DComEX/dcomex-framework --tag dcomex
$ docker run -it dcomex bash
inside the container run
$ cd /src/tests/units
$ ./run.sh
$ cd /src/tests/validation/inference_heat/
$ OMP_NUM_THREADS=1 ./run_inference.py --num-cores 12 --num-samples 200
To run one Msolve simulation
$ bio 1 2 3
1.0851841319006034E-05
$ bio -h
Usage: bio [-v] [-c] [-s] k1 mu time
MSolve simulation of tumor growth.
Positional arguments:
k1 The growth rate of the tumor, in units of 1/second.
mu The shear modulus of the tumor, in units of kPa.
time The timestep to use in the simulation, in units of days.
Options:
-v Print verbose output during the simulation.
-c Output the MSolve configuration file and exit.
-s Use a surrogate instead of the full simulation.
-h Display this help message and exit.
Environment Variables:
mph The path to the mesh of the simulated domain,
in .mph format.
Returns:
The volume of the tumor, in units of cubic millimeters.
The online documentation is at ReadTheDocs.
Install korali, integration tools, and Msolve application
$ make 'USER = 1' 'PREFIX = $(HOME)/.local' lkorali
$ make 'USER = 1' 'PREFIX = $(HOME)/.local' lbin lib
$ make 'USER = 1' 'PREFIX = $(HOME)/.local' lmsolve
Run examples
$ python examples/graph/korali0.py
log evidence: -2.490052761730272
1.0851841319006034E-05
$ bio -v 1 2 3
1.0851841319006034E-05
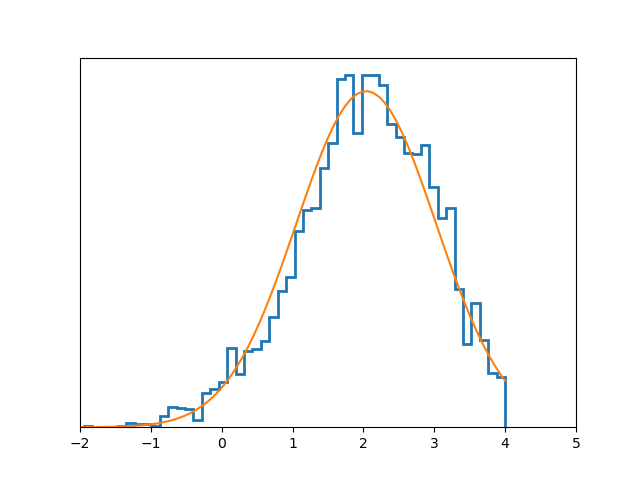
$ python examples/graph/analitical.py
graph.metropolis: accept = 0.724
graph.metropolis: accept = 0.684
graph.metropolis: accept = 0.7068
Open analitical.vis.png file
Running on Piz Daint
To replicate CI runs manually it is possible to pull the containers by logging in to Piz Daint and execute the commands
$ module load sarus
$ sarus pull IMAGE_NAME
$ srun --pty -C gpu -A GROUP_ID -N1 -n1 sarus run --mpi --tty IMAGE_NAME bash
This will drop you to a shell on the compute node inside the
container. From there you can replicate running the commands as in
ci/prototype.yml. It is important to make sure that the container
image names match the naming */public/*, i.e. they must reside in a
directory named public, only then anonymous access is possible.
- bin: integration utilities
- CI: definition of containerised build, test and deployment pipelines via CI/CD at CSCS
- docs: the source of the documentation
- examples: tutorials and examples
- graph.py: sample Bayesian graphs
- follow.py: trace function evalution and detect loops
- kahan.py: Kahan summation or compensated summation algorithms
- korali: a directory to build korali
- tests: unit and integration tests
Application
- msolve: Msolve application
- integration: integration of Msolve and korali