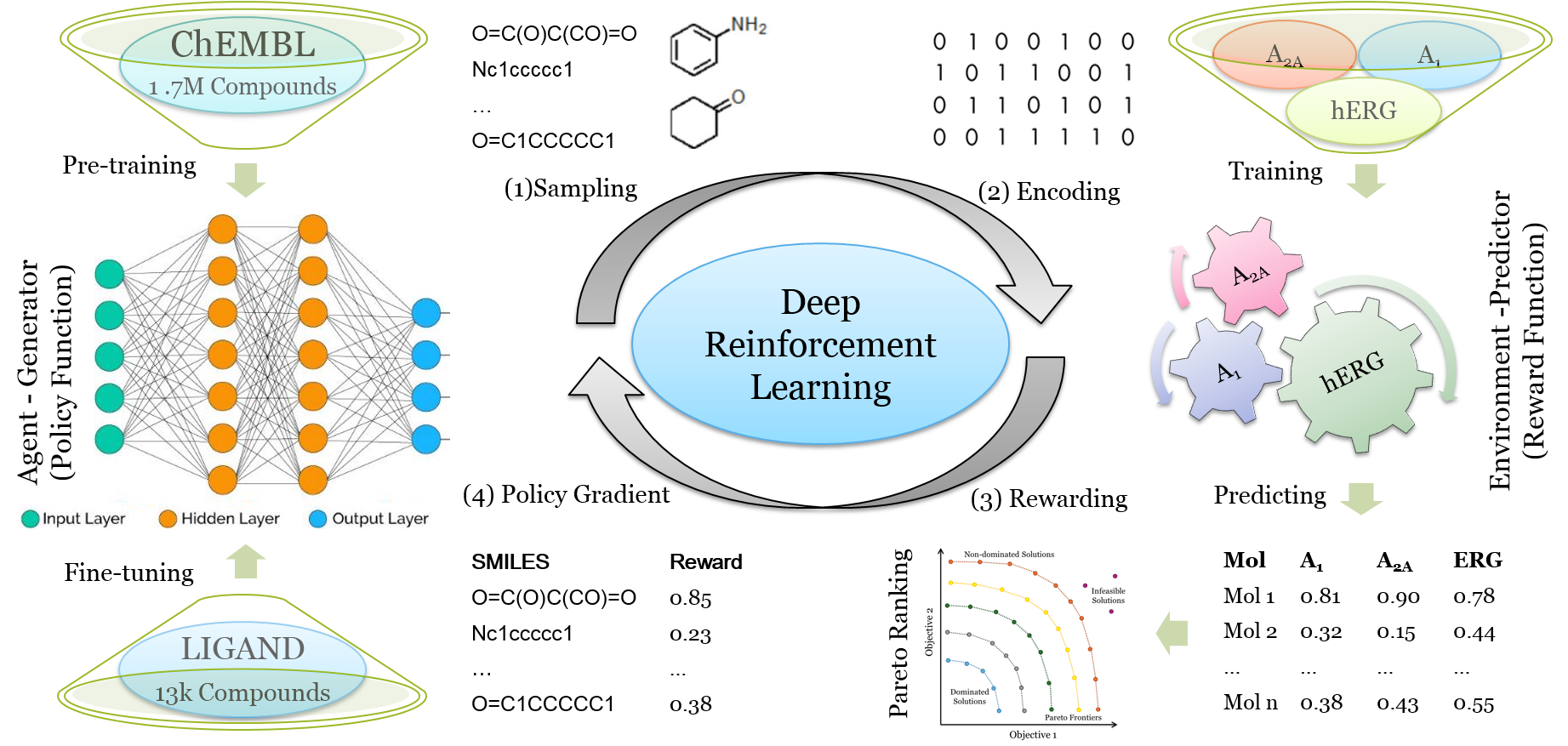
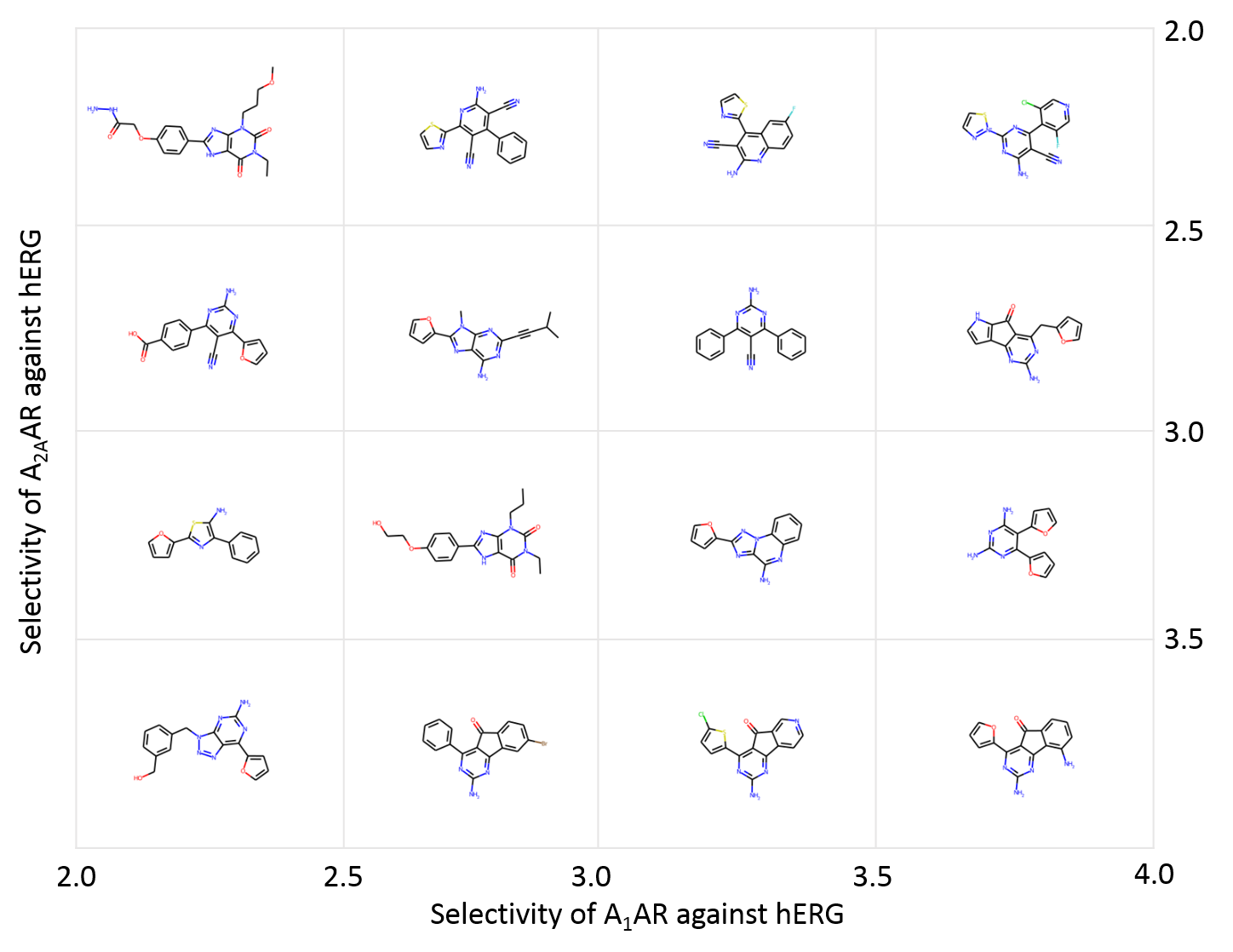
DrugEx v2 (Drug Explorer): De Novo Design of Drug Molecule by Pareto-based Multi-Objective Reinforcement Learning in Polypharmacology
By Xuhan Liu & Gerard J.P. van Westen, on March 8th 2021
Please see the LICENSE file for the license terms for the software. Basically it's free to academic users. If you do wish to sell the software or use it in a commercial product, then please contact us:
Xuhan Liu (First Author):
Gerard J.P. van Westen (Correspondent Author):
After achieving dramatic breakthrough in both image recognition, natural language processing and game playing, the deep learning methods are increasingly applied in drug discovery, e.g. recurrent neural networks (RNNs) have been shown to be an effective method to generate molecular libraries in drug discovery. We proposed a new method named DrugEx in the previous work that integrates an exploration strategy into RNN-based reinforcement learning to improve the diversity of the generated molecules. However, most current deep learning-based methods focus on a single target to generate drug-like active molecules whereas in reality drug molecules often interact with more than one target. This can have desired (polypharmacology) or undesired (toxicity) effects. Here, we extend our DrugEx algorithm with multi-objective optimization and generated drug molecules against more than one specific target (two adenosine receptors A1AR, A2AAR and hERG in this study). In our model, we applied an RNN as the agent and machine learning predictors as the environment, both of which were pre-trained in advance and then interplayed under reinforcement learning framework. And the idea of evolutionary algorithms was merged into our method such that crossover and mutation operations were implemented by the same deep learning model as the agent. During the training loop, the agent generates a batch of SMILES-based molecules. Subsequently scores for all objectives provided by the environment are used for constructing Pareto ranks of the generated molecules. Here, we adopted GPU acceleration to speed up the process of Pareto optimization. The final reward of each molecule is calculated based on the Pareto ranking with the ranking selection algorithm. The agent is trained under the guidance of the reward to make sure it can generate more desired molecules after convergence of the training process. We demonstrate generation of compounds with a diverse predicted selectivity profile toward multiple targets, offering the potential of high efficacy and lower toxicity.
Firstly, ensure that the version of your Python >= 3.7. We recommend Anaconda to manage the version of Python and installed packages.
Secondly, all the following packages are installed in your machine:
1. Numpy (version >= 1.19)
$ conda install numpy
2. Scikit-Learn (version >= 0.23)
$ conda install scikit-learn
3. Pandas (version >= 1.2.2)
$ conda install pandas
4. PyTorch (version == 1.6)
$ conda install pytorch torchvision cudatoolkit=x.x -c pytorch
Note: it depends on the GPU device and CUDA tookit
(x.x is the version of CUDA)
5. Matplotlib (version >= 2.0)
$ conda install matplotlib
6. RDKit (version >= 2020.03)
$ conda install -c rdkit rdkit
For designing the novel drug molecules with SMILES representation, you should do the following steps sequentially by running scripts:
-
dataset.py:
Preparing your dataset for pre-training and fine-tuning the RNN model as initial states of exploitation network and exploration network.
-
environ.py:
Training your predictor as the environment for providing the final reward for the action from the agent. The performance can also be evaluated through n-fold cross validation and independent test.
-
pretrainer.py:
Pre-training the RNN model as initialization of exploitation network acted as agent for molecule design. Fine-tuning the same RNN model as agent net which will be fixed as an pertubation to enlarge the diversity.
-
train.py:
Training the DrugEx model under the reinforcement learning framework. During the training process, both of the exploitation and exploitation network will be involved in the SMILES generation, and the mutation rate controls the contribution that exploration network makes.
-
sampler.py:
Finally, generating the SMILES format molecules with well-trained RNN model (pre-trained/fine-tuned model or DrugEx model).
-
figure.py:
It provides a variety of the methods to measure the performance of every step during the training process of DrugEx, and form the figure for results visualization.
In addition, this toolkit also provides some other scripts for definition of special data structures, model architectures and coefficient measurements, etc.
-
models/*.py:
It contains all of the deep learning models that possibly used in this project, including single/multiple fully-connected regression/classification models, RNN generative model and highway CNN classification model.
-
utils/vocab.py:
It defines some special data structures, such as vocabulary of SMILES tokens, molecule dataset, environment and some methods for SMILES checking. The statistical methods that extracting properties from generated molecules.
-
utils/metric.py:
The statistical methods that extracting properties from generated molecules.
-
utils/fingerprints.py:
There are a variety of chemical fingerprints calculations, such as ECFP, MACCS etc.
-
utils/modifier.py
It provides a variety of desirability function to normalize the scoring furntions. For more details, please check GuacaMol benchemark.
-
utils/objective.py
It provides the construction of different scoring functions, including similary score, chemical properties, QSAR modelling, etc. Moreoever, it can also integrate multiple objective into an environment to calculate reward for the agent.
-
utils/nsgaii.py
The implementation of non-dominate sorting and crowding distance algorithm (NSGAII). Importantly, we employ PyTorch to accelerate its performance and also modify the calculation of crowding distance with Tanimoto-distance.
-
utils/sacorer.py
The implementation of SA score to measure the synthezability score of each molecule. More details about SA score can be found here
We thank the following Git repositories that gave me a lot of inspirations: