This is the official repository for the paper "Rad-ReStruct: A Novel VQA Benchmark and Method for Structured Radiology Reporting", which published at MICCAI 2023. This repo includes our code as well as our structured reporting dataset.
We introduce Rad-ReStruct, a new benchmark dataset that provides fine-grained, hierarchically ordered annotations in the form of structured reports for X-Ray images. Our dataset provides structured labels for the images in the IU-XRay dataset available on OpenI [1]. We model the structured reporting task as hierarchical visual question answering (VQA) and propose hi-VQA, a novel method that considers prior context in the form of previously asked questions and answers for populating a structured radiology report. In this repo we provide the structured reports as well as the list of questions to fill this report for every image. Authors: Chantal Pellegrini, Matthias Keicher, Ege Özsoy, Nassir Navab
[1] Demner-Fushman, D., Antani, S., Simpson, M., & Thoma, G. R. (2012). Design and development of a multimodal biomedical information retrieval system. Journal of Computing Science and Engineering, 6(2), 168-177.
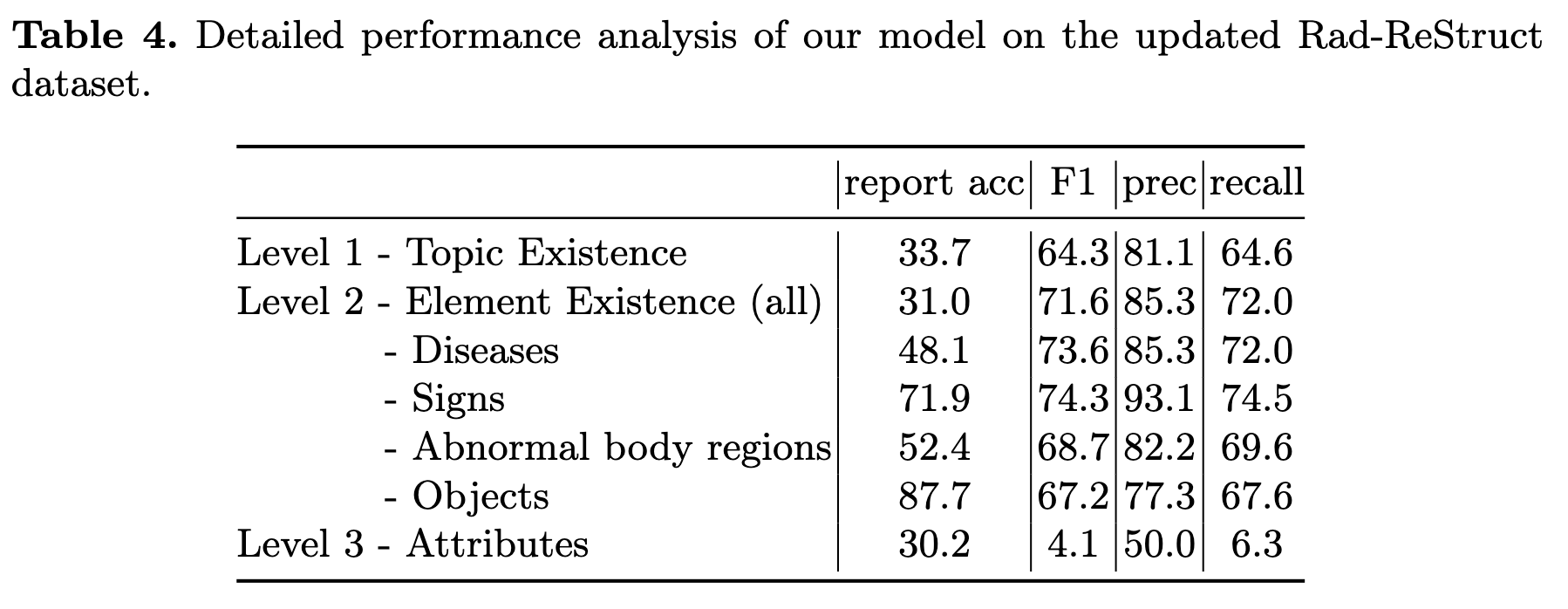
Since the publication of our paper, we have updated our dataset to fix some inconsistencies in the structured reports. The updated evaluation results on Rad-ReStruct are shown below. Please use these updated results when comparing to our method.
-
Clone this repository
git clone https://github.com/ChantalMP/Rad-ReStruct.git -
Install requirements:
conda create -n radrestruct_env python=3.9 conda activate radrestruct_env pip install -r requirements.txt -
Download data
Rad-ReStruct:
- download images from https://openi.nlm.nih.gov/imgs/collections/NLMCXR_png.tgz
- unpack and copy image files into 'data/radrestruct/images'
VQARad:
- download images as zip from https://osf.io/89kps/
- unpack and copy image files into 'data/vqarad/images'
- The dataset is available in the folder 'data/radrestruct'. The structured reports are in 'new_reports', the QA pairs are in '_qa_pairs'. _vectorized_answers contains on 2470-dim vector for every report in the dataset, where each dimension corresponds to one answer (see Report-based Evaluation). The report names correspond to the names of the original free-text reports (https://openi.nlm.nih.gov/imgs/collections/NLMCXR_reports.tgz).
- To load the dataset use the following code. This will load the dataset as VQA dataset, where one sample is an image together with a single question + history.
from data_utils.data_radrestruct import RadReStruct
# required parameters
parser = argparse.ArgumentParser(description="Finetune on RadReStruct")
parser.add_argument('--bert_model', type=str, required=False, default="zzxslp/RadBERT-RoBERTa-4m", help="pretrained question encoder weights")
parser.add_argument('--max_position_embeddings', type=int, required=False, default=458, help="max length of sequence")
parser.add_argument('--img_feat_size', type=int, required=False, default=14, help="dimension of last pooling layer of img encoder")
parser.add_argument('--num_question_tokens', type=int, required=False, default=20, help="number of tokens for question")
args = parser.parse_args()
args.num_image_tokens = args.img_feat_size ** 2
args.num_question_tokens = 458 - 3 - args.num_image_tokens
# load dataset
train_tfm, val_tfm = ... # image transforms, our transforms can be found in 'train_radrestruct.py' (line 100)
traindataset = RadReStruct(tfm=train_tfm, mode='train', args=args)
valdataset = RadReStruct(tfm=test_tfm, mode='val', args=args)
- For evaluation, we need to ensure consistency of the entire report, therefore we provide an evaluation dataset, where one sample is an image together with the entire structured report. To load this dataset use the following code:
from data_utils.data_radrestruct import RadReStructEval
# required parameters
...
# load dataset
val_tfm = ... # image transforms, our transforms can be found in 'evaluate_radrestruct.py' (line 100)
valdataset = RadReStructEval(tfm=test_tfm, mode='test', args=args, limit_data=2)
- To evaluate your model with our metrics, we provide an evaluator class, which can be used as follows:
from evaluation.evaluator_radrestruct import AutoregressiveEvaluator
preds = ...
evaluator = AutoregressiveEvaluator()
targets = get_targets_for_split('test')
acc, acc_report, f1, precision, recall, detailed_metrics = evaluator.evaluate(preds, targets)
-
The evaluator expects a 2470-dim binary prediction vector per report. Each element denotes if a certain answer to a question is set to true or false. The rows of this vector are defined in 'data/radrestruct/report_keys.json' and correspond to all possible paths in the report template. Answers to questions that are not predicted due to hierarchy can be set to 0. The evaluator will clean the prediction, setting the "no selection" option if no predictions are made and enforce consistency in the prediction, by setting lower-level question following a negative answer to no / no selection. Therefore, your predictions might change during evaluation.
-
We provide our code for predicting this vector using auto-regressive evaluation in 'evaluation/predict_autoregressive_VQA_radrestruct.py'. You can use this as reference, but will need to adapt it for your approach.
training: python -m train.train_radrestruct --run_name "train_radrestruct" --classifier_dropout 0.2 --acc_grad_batches 2 --lr 1e-5 --epochs 200 --batch_size 32 --progressive --mixed_precision
evaluation: python -m evaluation.evaluate_radrestruct --run_name "eval_radrestruct" --match_instances --batch_size 1 --use_pretrained --model_dir "<WEIGHT_PATH>" --progressive --mixed_precision
To use our pretrained model, download the weights from release v.1.0.0 and set WEIGHT_PATH pointing to the weight file.
training: python -m train.train_vqarad --run_name "train_vqarad" --acc_grad_batches 2 --lr 5e-5 --epochs 200 --batch_size 32 --progressive --mixed_precision
evaluation: python -m evaluation.evaluate_autoregressive_vqarad --run_name "eval_vqarad" --batch_size 1 --use_pretrained --model_dir "<WEIGHT_PATH>" --progressive --mixed_precision
To use our pretrained model, download the weights from release v.1.0.0 and set WEIGHT_PATH pointing to the weight file.
This model is intended to be used solely for (I) future research on visual-language processing and (II) reproducibility of the experimental results reported in the reference paper.
When using our dataset please cite both our paper and the original IU-XRay paper:
@article{pellegrini2023radrestruct,
title={Rad-ReStruct: A Novel VQA Benchmark and Method for Structured Radiology Reporting},
author={Pellegrini, Chantal and Keicher, Matthias and {\"O}zsoy, Ege and Navab, Nassir},
journal={TODO},
year={2023}
}
@article{demner2012design,
title={Design and development of a multimodal biomedical information retrieval system},
author={Demner-Fushman, Dina and Antani, Sameer and Simpson, Matthew and Thoma, George R},
journal={Journal of Computing Science and Engineering},
volume={6},
number={2},
pages={168--177},
year={2012},
publisher={Demner-Fushman Dina; Antani Sameer; Simpson Matthew; Thoma George R.}
}