https://www.nature.com/articles/s41467-023-41454-9
This repository contains the PyTorch implementation of A pharmacophore-guided deep learning approach for bioactive molecular generation.
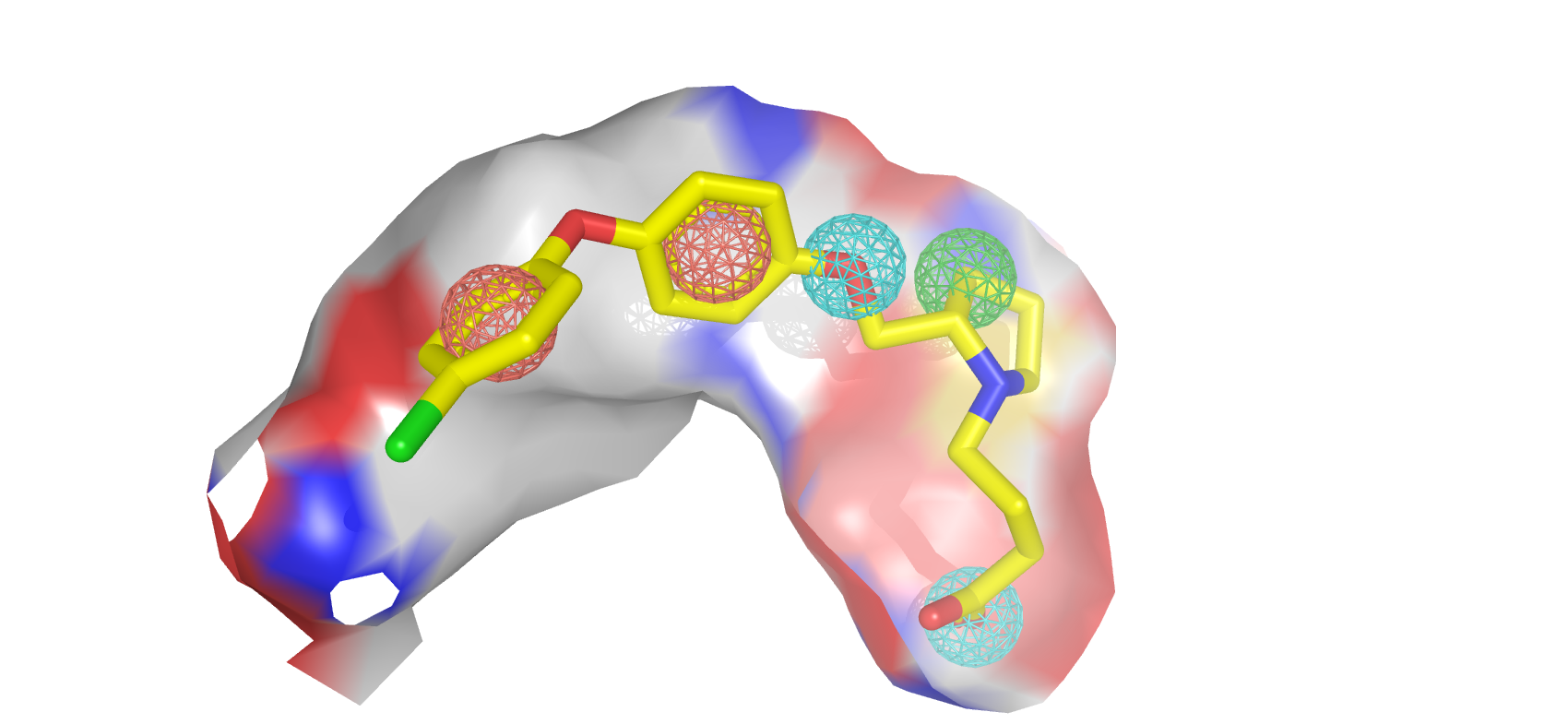
Through the guidance of pharmacophore, PGMG provides a flexible strategy to generate bioactive molecules with structural diversity in various scenarios using a trained variational autoencoder.
Update on 02/05/2023: The PGMG web server is now available! Experience PGMG without having to perform any deployments on your own.
PGMG aims to provide a flexible strategy to generate bioactive molecules with structural diversity in various scenarios, especially when the activity data is scarce.
PGMG only requires a pharmacophore hypothesis as input. The hypothesis can be constructed using only a few ligands or the structure of the receptor or the ligand-receptor complex. The pharmacophore hypothesis will be transformed into a weighted complete graph based on the shortest-path distance and feed into the model. The model will then rapidly generate a large number of molecules that satisfy the conditions.
- python==3.8
- torch==1.12.1+cu102
- rdkit==2022.09.1
- dgl-cuda10.2==0.9.1
- fairseq==0.10.2
- numpy==1.23.5
- pandas==1.5.2
- tqdm==4.64.1
- einops==0.6.0
If you encounter
RuntimeError: mat1 and mat2 shapes cannot be multiplied, please check the version offairseqfirst. It should be10.2.
We recommend using conda to manage the environment.
conda env create -f environment.ymlThe training process with default parameters requires a GPU card with at least 10GB of memory.
Run train_chembl_baseline.py using the following command:
CUDA_VISIBLE_DEVICES=<gpu_num> python train_chembl_baseline.py <output_dir> --show_progressbar- the
gpu_numindicates which gpu you want to run the code - the
output_diris the directory you want to store the trained model
Other configurations need to be changed inside train_chembl_baseline.py, including model settings and the data directory.
It takes about 70 hours to run the training script with default parameters using a single 2080Ti.
First of all, you need some pharmacophore hypotheses. A pharmacophore is defined as a set of chemical features and their spatial information that is necessary for a drug to bind to a target and there are many ways to acquire one.
If you have a biochemistry background, we strongly encourage you to build it yourself by stacking active ligands or analyzing the receptor structure. There are also many tools available. And you can always adjust the input hypothesis according to the results.
Apart from building it yourself, you can also acquire them by searching the literature or just randomly sampling 3-6 pharmacophore elements from a reference ligand to build some hypotheses and filtering the generated molecules afterwards.
The pharmacophore hypotheses need to be converted to a fully-connected graph and should be provided in one of the two formats:
- the
.pospformat where the type of the pharmacophore points and the 3d positions are provided, seedata/phar_demo2.pospfor example. - the
.edgepformat where the type of the pharmacophore points and the shortest-path-based distances between each point are provided, seedata/phar_demo1.edgepfor example.
Pharmacophore types supported by default:
- AROM: aromatic ring
- POSC: cation
- HACC: hydrogen bond acceptor
- HDON: hydrogen bond donor
- HYBL: hydrophobic group (ring)
- LHYBL: hydrophobic group (non-ring)
The 3d position in .posp files will first be used to calculate the Euclidean distances between each point and then the distances will be mapped to the shortest-path-based distances.
See the Supplemental Information of our paper for detailed descriptions.
How to calculate shortest-path distances:
The shortest path between pharmacophore point a and b is calculated as
0.2*N, where N is number of heavy atoms in the pharmacophore point.
For example, the shortest-path distances between A-B and A-C in the picture below can be calculated as:
Use the generate.py to generate molecules.
usage:
python generate.py [-h] [--n_mol N_MOL] [--device DEVICE] [--filter] [--batch_size BATCH_SIZE] [--seed SEED] input_path output_dir model_path tokenizer_path
positional arguments:
input_path the input file path. If it is a directory, then every file ends with `.edgep` or `.posp` will be processed
output_dir the output directory
model_path the weights file (xxx.pth)
tokenizer_path the saved tokenizer (tokenizer.pkl)
optional arguments:
-h, --help show this help message and exit
--n_mol N_MOL number of generated molecules for each pharmacophore file
--device DEVICE `cpu` or `cuda`, default:'cpu'
--filter whether to save only the unique valid molecules
--batch_size BATCH_SIZE
--seed SEED
The output is a .txt file containing the generated SMILES. It takes about 30 seconds to generate 10,000 molecules using a single 2080Ti, and about 10 minutes if using CPUs.
To run generation on the demo input:
python generate.py data/phar_demo1.edgep demo_result/ weights/chembl_fold0_epoch32.pth weights/tokenizer.pkl --filter --device cpuWe provide the weights file acquired using train_chembl_baseline.py in the release page. Please unzip it in the root directory.
The current model only support a maximum of 8 pharmacophore points in a single hypotheis. If you want to increase the maximum number, a possible way is to re-train the model with increased number of randomly selected pharmacophore elements and a larger MAX_NUM_PP_GRAPHS.
Use get_match_score(smiles,dgl_graph) in utils.match_eval to calculate the match score between molecules and pharmacophores.
For example:
from pathlib import Path
from utils.file_utils import load_phar_file
from utils.match_eval import get_match_score
smiles_list = ['Cc1ccc(C(=O)Nc2c(C(N)=O)sc3ncccc23)o1', 'O=C(NC1CCCCC1)c1cc2c(nc(O)c3ccccc32)s1']
file_path = Path('data/phar_demo1.edgep')
dgl_graph = load_phar_file(file_path)
dgl_graphs = [dgl_graph, dgl_graph]
match_scores = get_match_score(dgl_graphs, smiles_list, n_workers=8, timeout=20) # [0.67, 1.0]This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License.
For commercial use, please contact [email protected].