A compute graph for loading and transforming data for OWID.
Status: in production
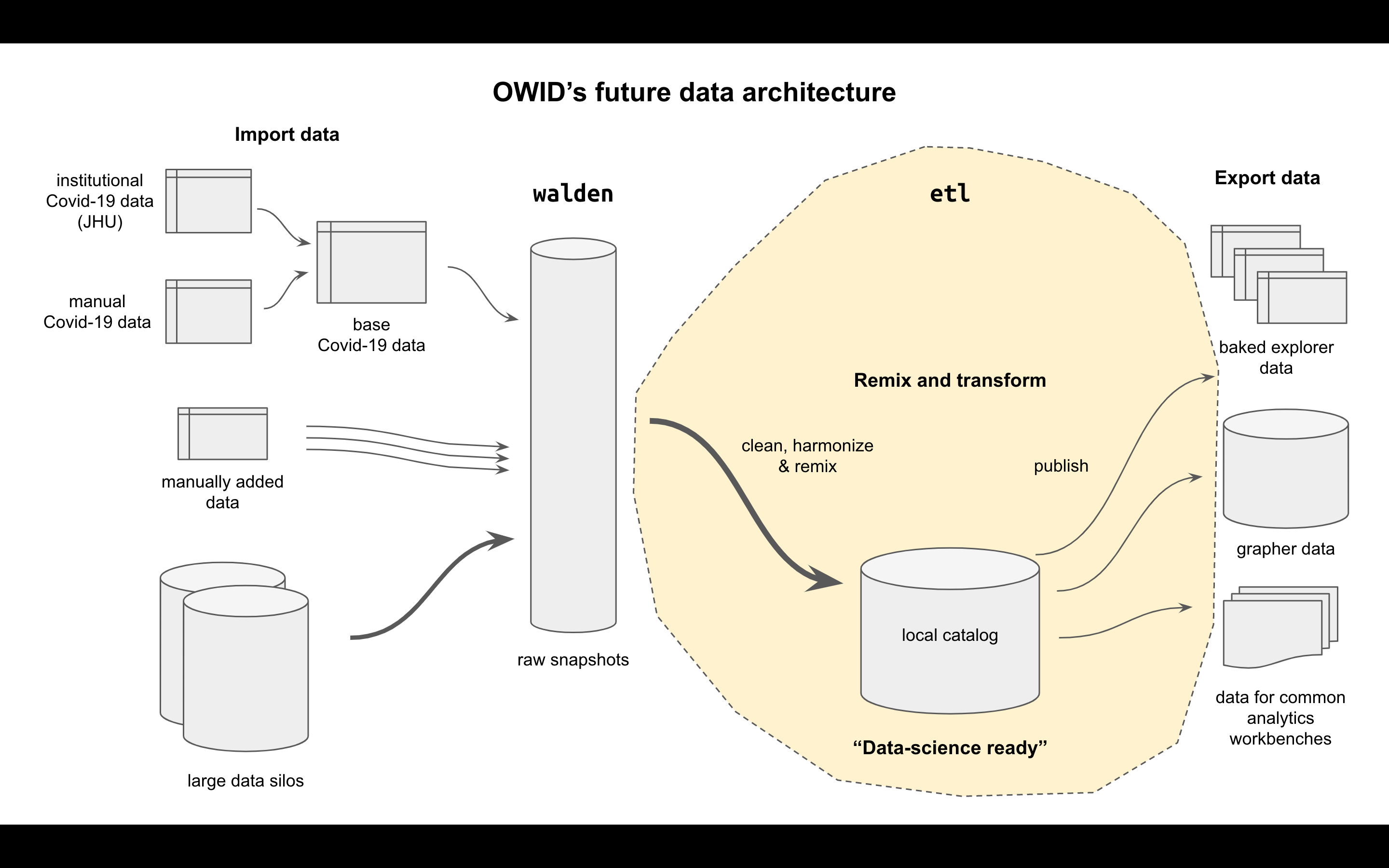
This project is the spiritual successor to importers, meant to eventually replace it. Its job is to assemble and republish OWID's data science catalog, the richest and easiest to use version of the datasets we can create. The catalog it creates is also meant to eventually replace Grapher's database as our source of truth for all data.
graph TB
upstream[Upstream data providers] -->|snapshot| walden[Walden snapshot]
upstream -->|publish| github
github[Datasets on Github] --> etl:::focus
walden --> etl(ETL steps)
etl -->|generate| catalog[Data catalog]
catalog -->|ingested| mysql
catalog -->|renders| explorers
catalog -->|published| api
mysql[Grapher MySQL] --> owid[Our World in Data site]
explorers --> owid
api --> jupyter[Jupyter notebooks]
classDef focus stroke-width:4px
You need to install the following:
- Python 3.9+. Guide for all platforms here. If you are using multiple different versions of Python on your machine, you may want to use
pyenvto manage them (instructions here). poetry, for managing dependencies, virtual envs, and packages. Installation guide here.make. You likely already have this, but otherwise it can be installed using your usual package manager (e.g. on Mac,brew install make).- MYSQL client (and Python dev headers). If you don't have this already,
- On Ubuntu:
sudo apt install python3.9-dev mysql-client default-libmysqlclient-dev build-essential - On Mac:
brew install mysql-client
- On Ubuntu:
- AWS CLI (
pip install awscli) and you should have an~/.aws/configfile configured (ask someone for credentials) so that you can upload to walden etc.
WSL users need to set up Linux executables only by including/editing the /etc/wsl.conf file.
- Run
sudo vi /etc/wsl.conf, copy this:
[interop]
appendWindowsPath = false
- Press
ESCand type:qwto save and exit.
We use poetry to manage the virtual environment for the project, and you'll typically do your work within that virtual environment.
- Run
git submodule update --init - Run
poetry install, which creates a virtual environment in.venvusingmake - Activate the virtual env with
poetry shell
To run all the checks and make sure you have everything set up correctly, try
make test
if make test fails report it in #data-architecture or #tech-issues. The etl is undergoing constant development so it may not be your local setup causing make test to fail and therefore shouldn't stop you progressing to the next step.
To run a subset of examples, you can try (for example)
poetry run etl covid
or
poetry run etl examples
These will generate files in ./data directory according to their recipes in ./etl/steps (with their dependencies defined in dag.yml).
Scripts in ./etl/steps/data/examples/examples/latest showcase basic functionality and can help you get started (mainly script_example.py).
You can also build all known data tables into the data/ folder with:
poetry run etl
However, processing all the datasets will take a long time and memory.
Note: poetry run runs commands from within the virtual environment. You can also activate it with poetry shell and then simply run etl ....
You can start an interactive walkthrough that will guide you through all the steps of creating a new dataset. Start it with
poetry run walkthrough walden
and then follow the instructions (two more steps are ran subsequently with poetry run walkthrough meadow and poetry run walkthrough garden).
In case you only want to get a better intuition of the process and see what files get generated, run
poetry run walkthrough walden --dummy-data
If you end up with error Address already in use, try to kill the process with kill -9 $(lsof -t -i tcp:8082) and run the command again.
Alternatively, these are the steps to create a data pipeline for a dataset called example_dataset, from an institution called
example_institution, with version YYYY-MM-DD (where this date tag can typically be the current date when the dataset
is being added to etl, or the date when the source data was released or updated):
- Activate the virtual environment (running
poetry shell). - Create a new branch in the
waldensubmodule. - Create an ingest script (e.g.
etl/vendor/walden/ingests/example_institution.py) to download the data from its original source and upload it as a new data snapshot into the S3waldenbucket. This step can also be done manually (although it is preferable to do it via script, to have a record of how the data was obtained, and to be able to repeat the process in the future, for instance if another version of the data is released). Keep in mind that, if there is additional metadata, it should also be ingested intowaldenas part of the snapshot. If the data is in a single file for which you have a download link, this script may not be required: you can add this link directly in the index file (see next point). There is guidance on how to upload towaldenmanually in thewaldenREADME. - Create an index file
etl/vendor/walden/index/example_institution/YYYY-MM-DD/example_dataset.jsonfor the new dataset. You can simply copy another existing index file and adapt its content. This can be done manually, or, alternatively, the ingest script can also write one (or multiple) index files. - Run
make testinwaldenand make changes to ensure the new files in the repository have the right style and structure. - Create a pull request to merge the new branch with the master branch in
walden. When getting started with theetlyou should request a code review from a more experiencedetluser. - Create a new branch in
etl. - Create a new
meadowstep file (e.g.etl/etl/steps/data/meadow/example_institution/YYYY-MM-DD/example_dataset.py). The step must contain arun(dest_dir)function that loads data from thewaldenbucket in S3 and creates a dataset (acatalog.Datasetobject) with one or more tables (catalog.Tableobjects) containing the raw data. Keep in mind that both the dataset and its table(s) should contain metadata. Additionally, all of the column names must be snake case before uploading tomeadow. There is a function in theowid.catalog.utilsmodule that will do this for you:tb = underscore_table(Table(full_df)). - Add the new meadow step to the dag, including its dependencies.
- Run
make testinetland ensure the step runs well. To run the step:etl data:https://meadow/example_institution/YYYY-MM-DD/example_dataset - Create a new garden step (e.g.
etl/etl/steps/data/garden/example_institution/YYYY-MM-DD/example_dataset.py). The step must contain arun(dest_dir)function that loads data from the lastmeadowstep, processes the data and creates a dataset with one or more tables and the necessary metadata. Country names must be harmonized (for which theharmonizetool ofetlcan be used). Add plenty of assertions and sanity checks to the step (if possible, compare the data with its previous version and check for abrupt changes). - Add the new garden step to the dag, including its dependencies.
- Run
make testinetland ensure the step runs well. - Create a new grapher step (e.g.
etl/etl/steps/data/grapher/example_institution/YYYY-MM-DD/example_dataset.py). The step must contain arun(dest_dir)function that loads data from the lastgardenstep, processes the data and creates a dataset with one or more tables and the necessary metadata. Add--grapherflags toetlcommand to upsert data into grapher database. To test the step, you can run it on the grapherstagingdatabase, or using a local grapher. - Create a pull request to merge the new branch with the master branch in
etl. At this point, some further editing of the step files may be required before merging the branch with master.
Please file any bugs or issues at https://github.com/owid/etl/issues. We are not currently seeking external contributions, but any member of the public should be able to run this codebase and recreate our catalog.
The etl project is the heart of OWID's future data architecture, containing all data transformations and publishing rules to take data from the raw snapshots kept in walden to the MySQL copy kept by grapher for publishing the OWID charts and site.
The ETL is the place where several key steps can be done:
- Syntactic harmonization: get data from institutional formats into a common OWID format with as few changes as possible
- Semantic harmonization: harmonize dimension fields like country, region, gender, and others to create OWID's reference data set
- Remixing: generating combined datasets and indicators, e.g. taking population from one dataset and using it to transforming another indicator into a per-capita version
- Republishing (OWID only): export OWID's reference data set for a variety of consumers
- Dependencies listed: all transformation steps and their dependencies are collected in a single file
dag.yml - URI style steps: all steps have a URI style format, e.g.
data:https://garden/who/2021-07-01/gho - Filenames by convention: We use convention to reduce the amount of config required
walden:https://<path>steps match data snapshots in the walden index at<path>, and download the snapshots locally when executeddata:https://<path>steps are defined by a Python or Jupyter notebook script inetl/steps/data/<path>.{py,ipynb}, and generate a new dataset in thedata/<path>folder when executed
- Data stored outside Git: unlike the importers repo, only tiny reference datasets and metadata is kept in git; all actual datasets are not stored and are instead regenerated on demand from the raw data in Walden
The core formats used are the Dataset and Table formats from owid-catalog-py.
- Dataset: a folder full of data files (e.g.
my_dataset/), with overall metadata inindex.json(e.g.my_dataset/index.json) - Table: a CSV or Feather file (e.g.
my_table.feather) with table and variable metadata in a.meta.jsonfile (e.g.my_table.meta.json)
Visit the owid-catalog-py project for more details on these formats or their Python API.
Walden is OWID's data store for bulk data snapshots. It consists of a data index stored in git, with the snapshots themselves stored in S3 and available over HTTPS.
Walden steps, when executed, find the matching data snapshot in Walden's index and ensure its files are downloaded, by calling ensure_downloaded() on it. Walden stores such locally cached files in ~/.owid/walden.
Each data step is defined by its output, it must create a new folder in data/ containing a dataset at the matching path. The name also indicates where the script to run for this step lives.
For example, suppose we have a step data:https://a/b/c. Then:
- The step must create when run a dataset at
data/a/b/c - It must have a Python script at
etl/steps/data/a/b/c.py, a module atetl/steps/data/a/b/c/__init__.py, or a Jupyter notebook atetl/steps/data/a/b/c.ipynb
Data steps can have any dependencies you like. The ETL system will make sure all dependencies are run before the script starts, but the script itself is responsible for finding and consuming those dependencies.
An empty step used only to mark a dependency on a Github repo, and trigger a rebuild of later steps whenever that repo changes. This is useful since Github is a good store for data that updates too frequently to be snapshotted into Walden, e.g. OWID's covid-19-data.
Example: github:https://owid/covid-19-data/master
The most recent commit hash of the given branch will be used to determine whether the data has been updated. This way, the ETL will be triggered to rebuild any downstream steps each time the data is changed.
NOTE: Github rate-limits unauthorized API requests to 60 per hour, so we should be sparing with the use of this step as it is implemented today.
A step used to mark dependencies on HTTPS resources. The path is interpreted as an HTTPS url, and a HEAD request is made against the URL and checked for its ETag. This can be used to trigger a rebuild each time the resource changes.
Example: etag:https://raw.githubusercontent.com/owid/covid-19-data/master/public/data/owid-covid-data.csv
A step to load a dataset from grapher channel into the grapher mysql database. This step is not defined in DAG, but is generated dynamically with the --grapher flag. The job of this script is to make the input dataset fit the constrained grapher data model where we only have the exact dimensions of year and entity id. The latter is the numeric id of the entity (usually the country) and the former can also be the number of days since a reference date. The dataset from grapher channel is re-fitted for the grapher datamodel and the rest is taken care of by the ETL library.
The .metadata.namespace field will be used to decide what namespace to upsert this data to (or whether to create this namespace if it does not exist) and the .metadata.short_name field will be used to decide what dataset to upsert to (again, this will be created if no dataset with this name in this namespace exists, otherwise the existing dataset will be upserted to). .metadata.sources will be used to upsert the entries for the sources table. Tables from the dataset will be upserted as an entry in the variables table.
The actual data values aren't upserted to mysql, but are loaded from respective parquet files in the grapher channel.
Firstly, edit dag.yml and add a new step under the steps section. Your step name should look like data:https://<path>, meaning it will generate a dataset in a folder at data/<path>. You should list as dependencies any ingredients your dataset will need to build.
To define a Python data step, create a new python module at etl/steps/data/<path>.py. Your module must define a run() method, here is the minimal one:
from owid.catalog import Dataset
def run(dest_dir: str) -> None:
ds = Dataset.create_empty(dest_dir)
ds.metadata.short_name = 'my_dataset'
ds.save()You can use make etl to rebuild everything, including your new table. Or you can run:
.venv/bin/etl data:https://<path>
just to run your step alone.
A Jupyter notebook step is nearly identical to the Python step above, but instead of a Python module you create a new notebook at etl/steps/data/<path>.ipynb.
Run make lab to start the Juypter Lab environment, then navigate to the folder you want, start a new notebook and rename it to match name in <path>.
Your notebook must contain a cell of parameters, containing dest_dir, like this:
dest_dir = '/tmp/my_dataset'If we tag this cell correctly, at runtime dest_dir will get filled in by the ETL system. To tag the cell, click on the cell, then the cog on the top right of the Jupyter interface. Add the tag parameters to the cell.
Add a second cell containing the minimal code to create a dataset:
from owid.catalog import Dataset
ds = Dataset.create_empty(dest_dir)
ds.metadata.short_name = 'my_dataset'
ds.save()
Then run make etl to check that it's working correctly before fleshing out your ETL step.
Frequently used patterns are provided in the etl/steps/data/examples folder. You can run them with
etl examples --force
or use them as starting templates for your own steps.
An interactive country harmonizing tool is available, which can generate a country mapping file.
.venv/bin/harmonize <path/to/input.feather> <country-field> <path/to/output.mapping.json>
Most of the steps have private versions with -private suffix (e.g. data-private:https://..., walden-private:https://...) that remember and enforce a dataset level is_public flag.
When publishing, the index is public, but tables in the index that are private are only available over s3 with appropriate credentials. owid-catalog-py is also updated to support these private datasets and supports fetching them over s3 transparently.
It is possible to upload private data to walden so that only those with S3 credentials would be able to download it.
from owid.walden import Dataset
local_file = 'private_test.csv'
metadata = {
'name': 'private_test',
'short_name': 'private_test',
'description': 'testing private walden data',
'source_name': 'test',
'url': 'test',
'license_url': 'test',
'date_accessed': '2022-02-07',
'file_extension': 'csv',
'namespace': 'private',
'publication_year': 2021,
}
# upload the local file to Walden's cache
dataset = Dataset.copy_and_create(local_file, metadata)
# upload it as private file to S3
url = dataset.upload(public=False)
# update PUBLIC walden index with metadata
dataset.save()--private flag rebuilds everything including private datasets
etl --private
reindex
publish --private
CRON job is running on a server every 5 minutes looking for changes on master on the etl repo. If there are changes it will run make publish which is equivalent to running
1. `etl` (build/rebuild anything that’s missing/out of date)
2. `reindex` (generate a catalog index in `data/` for each channel)
3. `publish` (rsync the `data/` folder to an s3 bucket s3:https://owid-catalog/)
Then the s3 bucket has a CloudFlare proxy on top (https://catalog.ourworldindata.org/). If you use the owid-catalog-py project from Python and call find() or find_one() you will be doing HTTP requests against the static files in the catalog.
Datasets from our production grapher database can be backported to ETL catalog. The first step is getting them to Walden using
bulk_backport
(specify --limit to make it process only a subset of datasets). It goes through all public datasets with at least one variable used in a chart and uploads them to Walden catalog (or skip them if they're already there and have the same checksum). If you set --skip-upload flag, it will only persist the datasets locally. You need S3 credentials to upload them to Walden.
Note that you still have to commit those updates to Walden index, otherwise others won't be able to rerun the ETL. (If you don't commit them, running etl and publish steps will still work for you in your local repository, but not for others).
Backported walden datasets can be processed with ETL using
etl --backport
(or etl backport --backport). This will transform original datasets from long format to wide format, optimize their data types, convert metadata and add them to the catalog. Then you can run publish to publish the datasets as usual.
Fastback is a service that polls grapher database for dataset updates and backports them as fast as possible. When it detects a change it will:
- Run
backportto get the backported dataset to local Walden catalog (without uploading to S3) - Run ETL on the backported dataset
- Publish new ETL catalog to S3
All these steps have been optimized to run in a few seconds (except of huge datasets) and make them available through data-api.
Internal OWID staff only
We have a staging environment for ETL that runs automatically on every push to staging branch. You can push directly to staging branch as it's quite unlikely someone else would be using it at the same time. Don't bother with creating pull requests for merging to staging. Pushing to staging goes something like this:
# rebase your branch on master
git checkout mybranch
git rebase origin/master
# reset staging to your branch
git checkout staging
git reset --hard mybranch
# push staging and let it execute automatically
git pushIf the ETL fails on staging, you'll see it in #analytics-errors channel on Slack. You can check logs in /tmp/etl-staging.log on our owid-analytics server or ssh into it and rerun the ETL manually:
ssh owid-analytics
cd etl-staging
.venv/bin/etl garden explorersInternal OWID staff only
Grapher step writes metadata to mysql and stores data as parquet files in the grapher channel. Admin still uses two ways of loading data - from table data_values for manually uploaded datasets and now from bucket owid-catalog in S3 (proxied by https://owid-catalog.nyc3.digitaloceanspaces.com/) for ETL datasets.
During local development your data isn't yet in S3 catalog, but is stored in your local catalog. You have to add the following env variable to owid-grapher/.env
CATALOG_PATH=/path/to/etl/data
Internal OWID staff only
Running ENV=.env.prod etl mystep --grapher is not enough to get your dataset into production. Data values in parquet format also need to be published into S3 bucket owid-catalog. That happens automatically when you merge to master branch. Then you have to run ENV=.env.prod etl mystep --grapher as usual (this is gonna be automated soon). If you are only making changes to the metadata and want to test it out, you don't have to merge it and just run ENV=.env.prod etl mystep --grapher locally.