2019-03-26
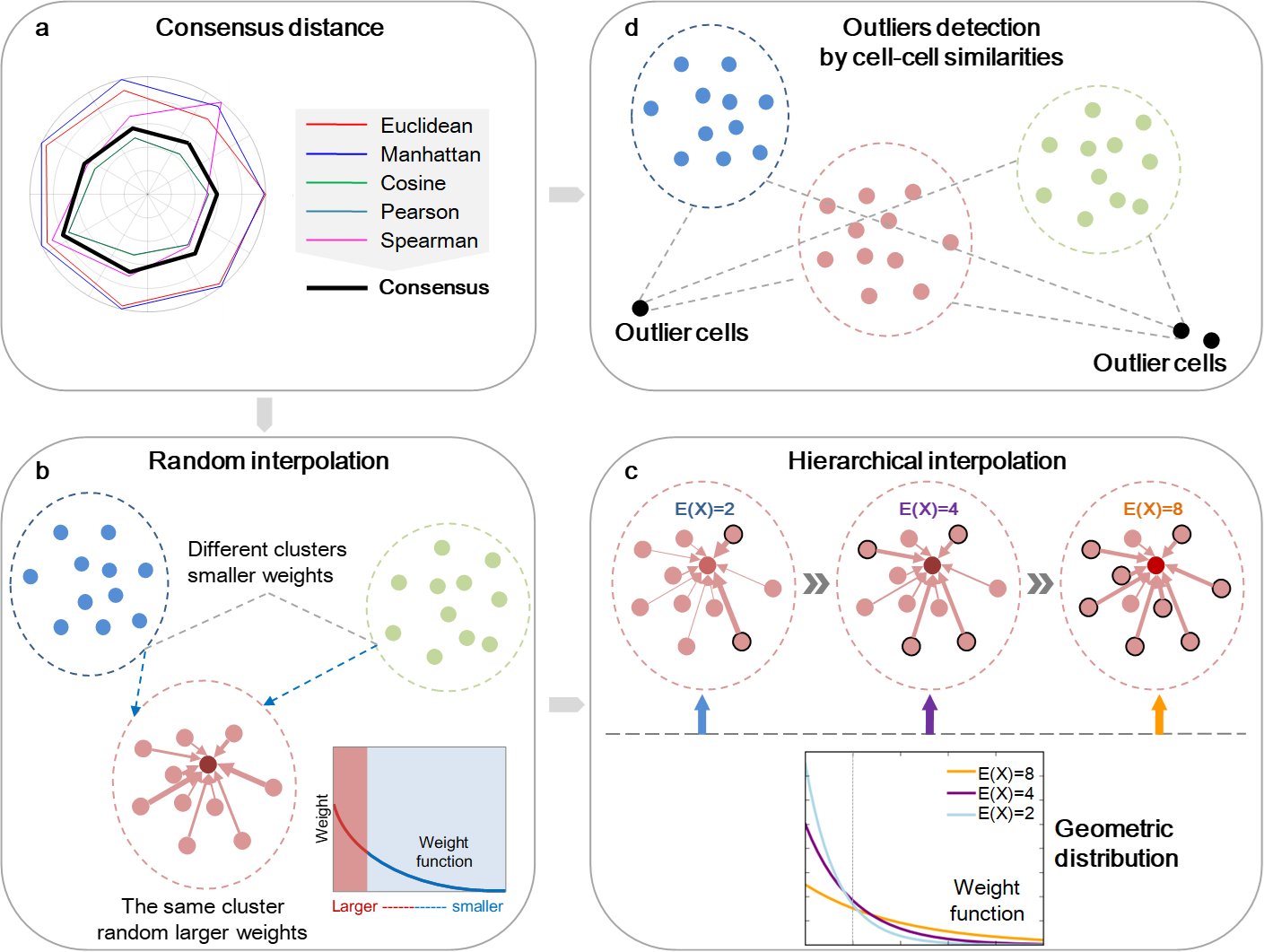
scHinter is a Matlab package for imputing dropout events for scRNA-seq with special emphasis on data with limited sample size. scHinter consists of three modules (Figure 1), incorporates a voting-based consensus distance and leverages the synthetic minority over-sampling technique for random interpolation. A hierarchical framework is also embedded in scHinter to increase the reliability of the imputation for small samples. The imputed expression matrix from scHinter can be used for as inputs for other existing scRNA-seq pipelines or tools for downstream analyses, such as cell type clustering, dimension reduction, and visualization.
(a) The voting-based consensus distance assembles several widely used distances into one consensus solution. (b) The random interpolation adopts the SMOTE (Synthetic Minority Over-sampling Technique) oversampling strategy and assigns random weights to cells according to the consensus distance. (c) The hierarchical framework was performed in a multi-layer manner, which gradually adds more cells for random interpolation and employs the probability density curves of geometric distribution with different mathematical expectations to weight cells under each layer.
The input to scHinter is matrix of gene expression. The rows correspond to cells and the columns correspond to genes. In this study, we will use the human tissues cells data from[1] as example.
clear;clc;
addpath('...\code');
load('...\matlab.mat');
scHinter firstly normalized the expression matrix by the library size of each cell, so that each cell will have an equal transcript count. This effectively eliminates cell size as a signal in the measurement for the purposes of constructing the affinity matrix and thus the resulting weighted neighborhood is not biased by cell size.
libsize = sum(data,2); data = bsxfun(@rdivide, data, libsize) * median(libsize);
scHinter calculate Cell-cell similarities learned by a voting-based consensus distance metric. Here, five distance measures were used to rank similarity between cells, including Euclidean distance, Manhattan distance, cosine distance, Pearson correlation coefficient, and Spearman correlation coefficient[2][3][4]. Parameter w is a weight for vote on the importance of different measures.
[TOT_rank]=dist_consensus(data,w);
scHinter implements random interpolation inspired by the oversampling strategy of SMOTE[5], which allows obtaining more data for classes with small sample size by oversampling to mitigate the imbalance of sample number in different classes. The expression matrix is updated after random interpolation. The corresponding parameter details can be referred to the comments in the code and supplementary materials of the article.
data_new=rdint(data,TOT_rank,sgm,m,thres,f)
Random interpolation was embedded under a hierarchical framework (Supplementary Fig. 1c), where multiple layers of interpolation are iteratively conducted by gradually including more cells as the most similar cells to the target cell.
data_new=data;
for qq=1:length(ex)
x=1:size(data,1);
p=1/ex(qq);
sgm=f*(1-p).^(x-1).*p;
data_new=rdint(data_new,TOT_rank,sgm,m,thres,f)
end
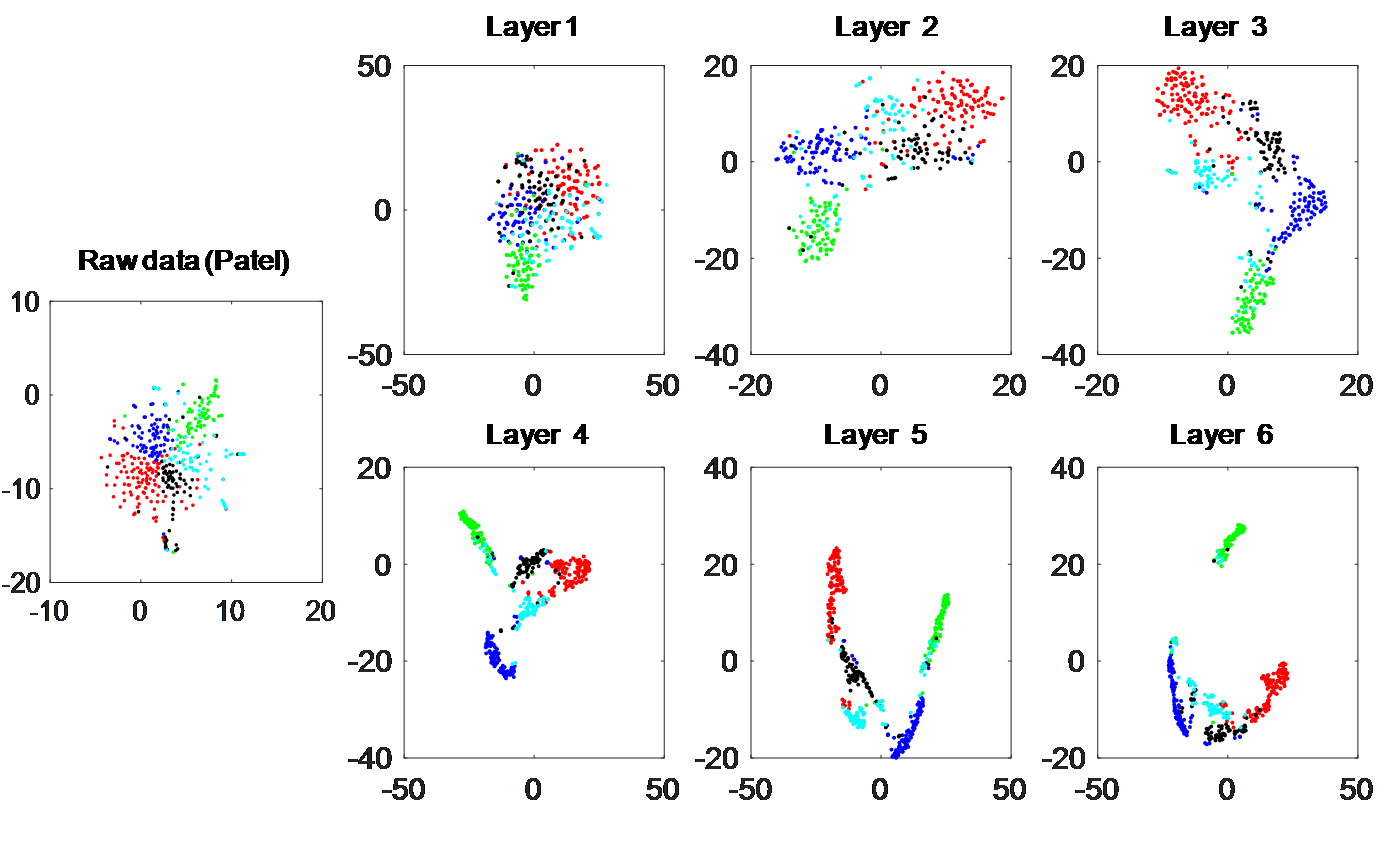
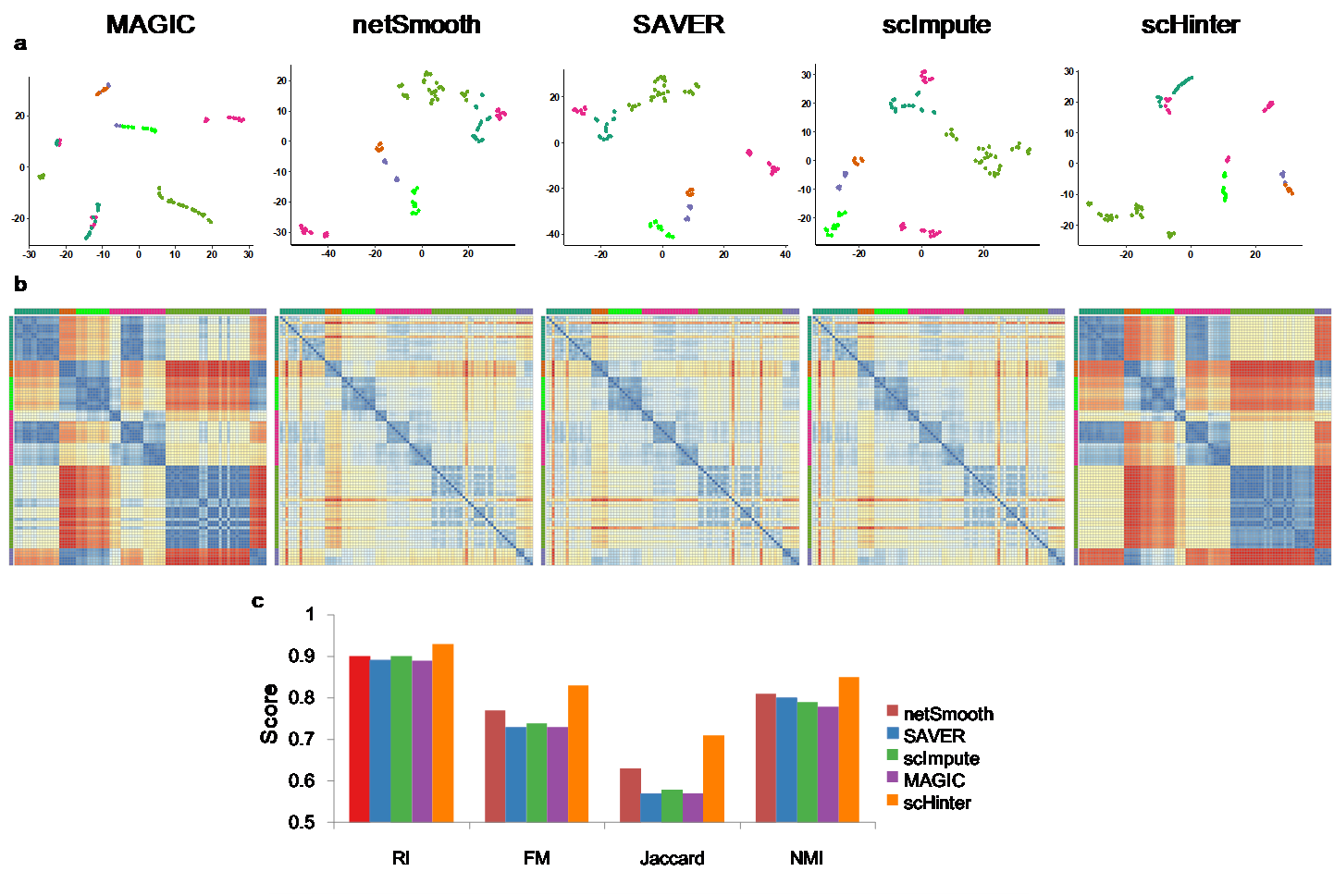
Figure 2 shows the t-SNE distribution effect of the data in the example after hierarchical interpolation. In addition, Figure 3 shows the distribution of each data in the article after imputing, including the two-dimensional distribution, heat map and bar chart of external performance of clustering metrics. Other performance results are described in detail in the article.
Figure 2. Illustration of the dynamic interpolation process under the hierarchical framework of scHinter.
Patel data was used as an example and the maximum number of layers is set at 6 according to the average cell number in this dataset (86≈2^6). t-SNE visualization was applied on the raw gene-cell expression matrix or the matrix after each round of interpolation to show differences among cell populations.
(a) t-SNE visualization on the imputed matrix from each individual tool. (b) Heatmaps for similarities learned from the data by Euclidean distances. The scales in relative units denote the similarity. Cells with the same cell type (annotated by the colored axes) are grouped together. (c) SC3 clustering accuracy on the imputed gene-cell expression matrix according to four performance metrics, including RI (Rand Index), FM (Fowlkes and Mallows Index), Jaccard, and NMI (Normalized Mutual Information). SC3 clustering is repeated for 10 times and each bar indicates the average performance.
If you are using scHinter, please cite: Ye P, Ye W, Ye C, Li S, Ye L, Ji G and Wu X* (2020) scHinter: Imputing dropout events for single-cell RNA-seq data with limited sample size, Bioinformatics, 36, 789–797.
[1] A. P. Patel et al., “NIH Public Access,” Science (80-. )., vol. 344, no. 6190, pp. 1396–1401, 2014.
[2] D. Aran et al., “Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage,” Nat. Immunol., vol. 20, no. February, 2019.
[3] Z. Ji and H. Ji, “TSCAN: Pseudo-time reconstruction and evaluation in single-cell RNA-seq analysis,” Nucleic Acids Res., vol. 44, no. 13, p. e117, 2016.
[4] V. Y. Kiselev, A. Yiu, and M. Hemberg, “scmap - A tool for unsupervised projection of single cell RNA-seq data,” bioRxiv, p. 150292, 2017.
[5] N. V. Chawla, K. W. Bowyer, L. O. Hall, and W. P. Kegelmeyer, “SMOTE: Synthetic minority over-sampling technique,” J. Artif. Intell. Res., vol. 16, pp. 321–357, 2002.