This repository contains scripts, data, metadata, and results to perform transcriptomic, genetic, and metabolic analysis to "Abies vs ozone's project".
In these directories you will find some analysis to answer these project's particular aims:
-
To quantify the secondary metabolite relative abundance in tolerant and damaged trees during two ozone concentration periods. Metabolomic analysis
-
To identify tolerance sacred fir's origins. Genomic analysis
-
To evaluate the tolerant and damaged trees differential expression in two ozone concentration periods. Transcriptomic analysis
+----- Abies_vs_ozone/
| +--1.-Sampling/
| +--2.-Metabolomics/
| +--3.-Genomics/
| +--4.-Transcriptomics/
| +--5.-INFO_PROJECT/
| +--README.md
README.md: There is a markdown file about this project. This file includes the repository's disposition.
1.-Sampling: There is a directory with coordinates and the samples' disposition in omics analisys.
2.-Metabolomics: There are some metabolite-generated analysis with gas chromatograph spectrum mass (GC-SM). Data from html files were loaded into tables. Subsequently, relative abundance was calculated. Finally, all the samples' values were compared using a barplot, ANOVA, and PCA analysis.
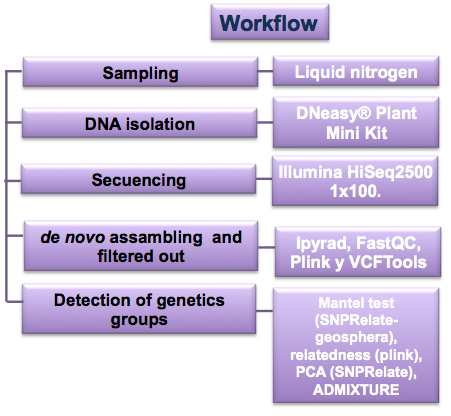
3.-Genomics: There are some genomic analysis from GBS sequencing. ipyRAD was used to assemble de novo, VCFTools and plink were used to make more specific filters. The relationship was calculated without multiple SNPs in the same loci. The mantel test, PCA, and admixture were performed in order to identify local origin samples.
4.-Transcriptomics: There are some transcriptomic analysis from RNAseq data. Samples were cut with Trimmomatic and they were mapped to a reference transcriptome with BWA. Rstudio allowed to evaluate differential expression between samples with edgeR and DESeq2. Subsequently, volcanoplot was performed to show overexpressed and underexpressed genes.
5.-INFO_PROJECT: There are some slide-shows, summaries, and final analysis.
To quantify secondary metabolite relative abundance in healthy and damaged trees during two ozone concenrations periods.
Check more information about this pipeline in README_metabolomics.
To see a short summary about the final metabolomics analysis click here.
Check more information about this pipeline in README_genomics.
To see a short summary about final genomic analysis click here.
Check more information about this pipeline in README_transcriptomics.
To see a short summary about final transcriptomic analysis click here.
Verónica Reyes Galindo
[email protected]