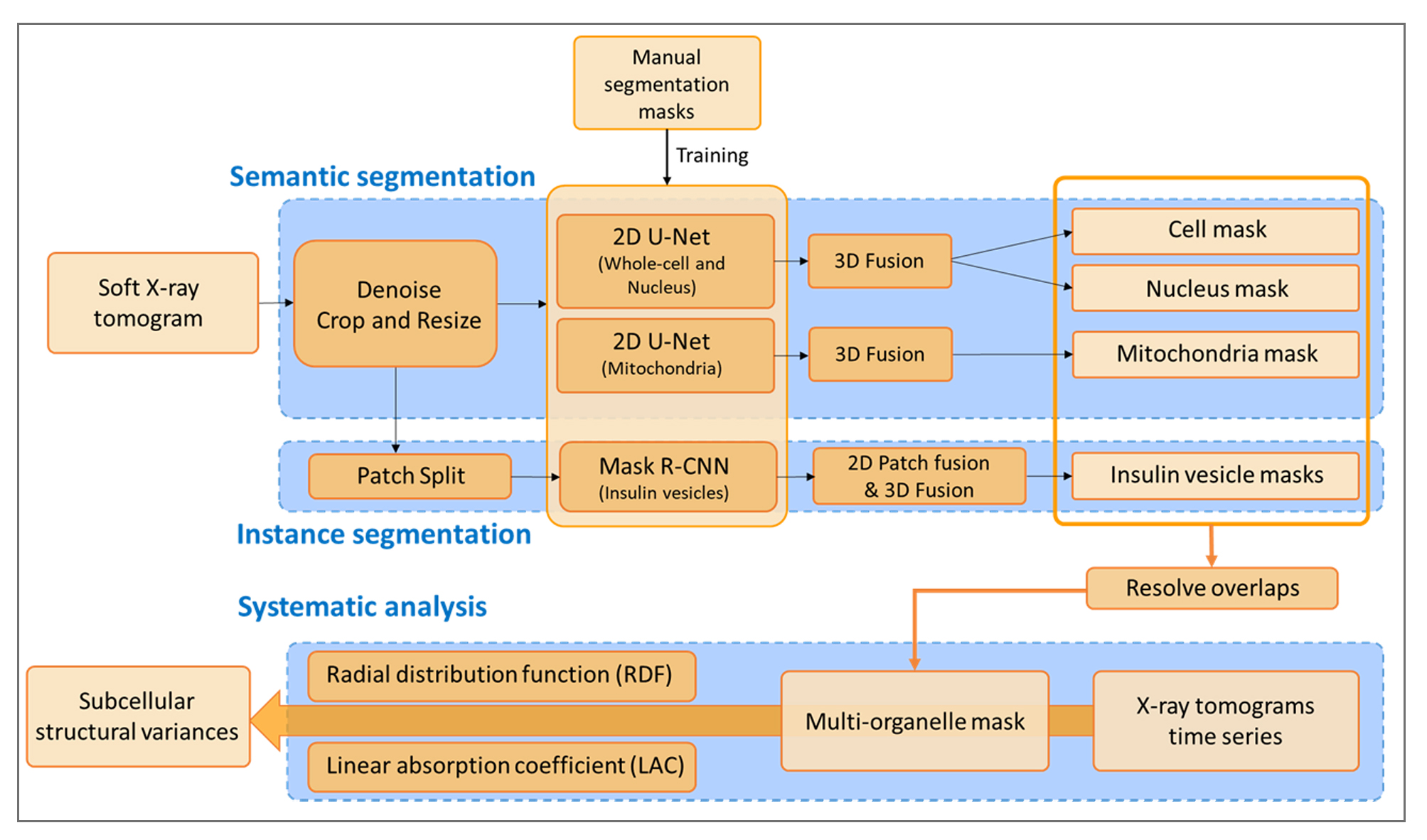
Official Implementation of Auto-Segmentation and Time-Dependent Systematic Analysis of Mesoscale Cellular Structure in β-Cells During Insulin Secretion.
- Python 3.7.3
- PyTorch 1.3.1
- torchvision 0.4.2
- Cuda version 10.0
git clone https://github.com/Xiangyi1996/Cell-Segmentation.git
cd Cell-Segmentation
mkdir logs
mkdir data
mkdir results
1. Download SXT dataset from PBC
Please go to PBC Dataset and download SXT images and manual labels, and put them under data folder.
# symlink the pascal dataset
cd data
ln -s /path_to_sxt_images/ ./image_xyz
ln -s /path_to_manual_labels/ ./mask_xyz
2. Download pretrained model Download the FS_mito and FS_mem_nu weights and put them under ./results folder.
# download the pretrained model
cd results
mkdir FS_mito
mkdir FS_mem_nu
# Test mitochondria model
sh script/test_mito.sh
# Test membrane and nuclear model
sh script/test_mem_nu.sh
There are some hyperparameters that you can adjust in the script.
python test/eval_mito.py --gpu 0 --exp FS_mito --num-workers 8 --batch-size 1 --num-classes 4 --test_idx 'iso'
PS: You can change the EXP in script to your pretrained model name. The above is just an example.
We list the performance w/o 3D fusion.
| Before 3D fusion | Membrane | Nucleus | Mito |
|---|---|---|---|
| 766_8 | 90.74 | 93.21 | 68.58 |
| 784_5 | 87.43 | 89.95 | 63.17 |
| 842_17 | 85.34 | 83.50 | 65.03 |
| mean | 87.84 | 88.89 | 65.59 |
| After 3D fusion | Membrane | Nucleus | Mito |
|---|---|---|---|
| 766_8 | 93.54 | 93.92 | 70.34 |
| 784_5 | 89.41 | 91.82 | 67.29 |
| 842_17 | 91.85 | 89.49 | 67.40 |
| mean | 91.60 | 91.74 | 68.34 |