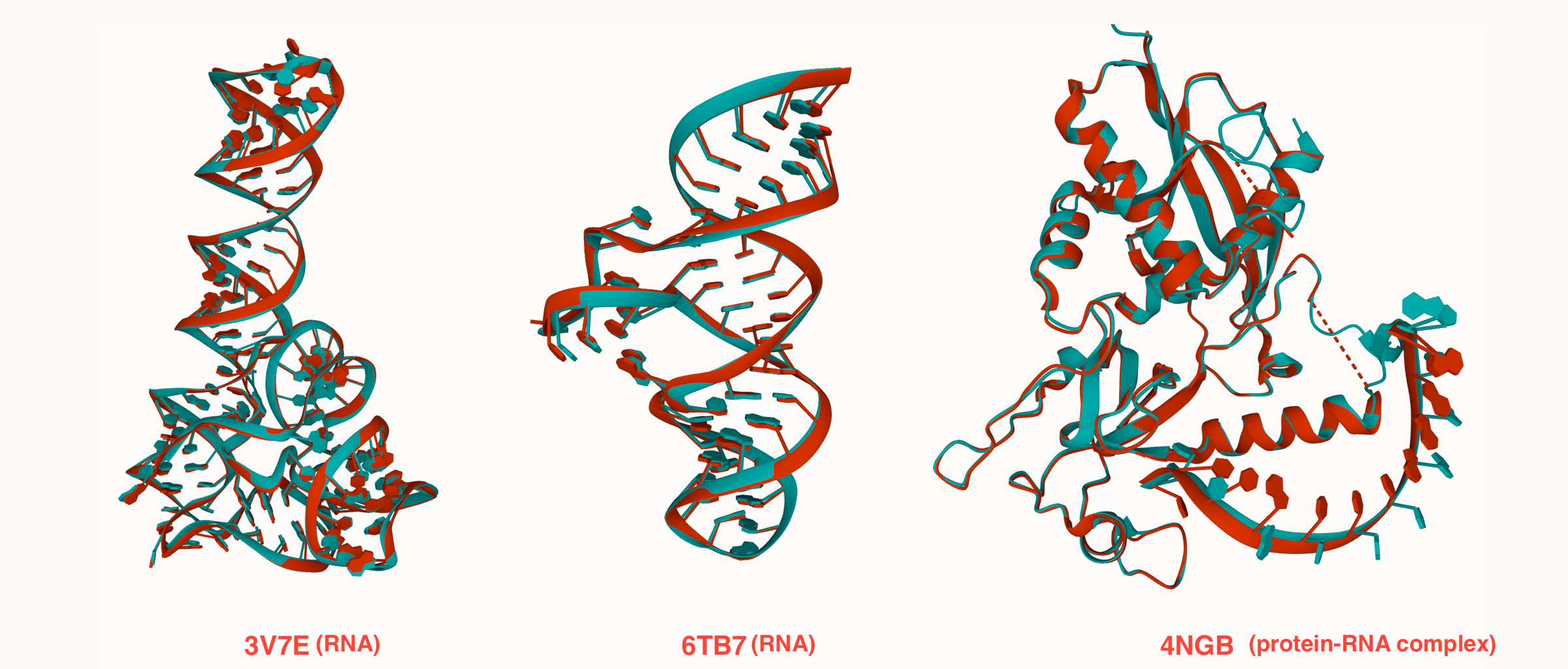
OpenComplex is an open-source platform for developing protein and RNA complex models.
Based on DeepMind's Alphafold 2 and AQ Laboratory's OpenFold, OpenComplex support almost all features from Alphafold 2 and OpenFold, and introduces the following new features:
- Reimplemented Alphafold-Multimer models.
- RNA and protein-RNA complex models with high precision.
- Kernel fusion and optimization on >=Ampere GPUs, brings 16%
We will release training results and pretrained parameters soon.
All Python dependencies are specified in environment.yml. For producing sequence
alignments, you'll also need kalign, the HH-suite,
and one of {jackhmmer, MMseqs2 (nightly build)}
installed on on your system.
Finally, some download scripts require aria2c and aws.
For convenience, we provide a script that installs Miniconda locally, creates a
conda virtual environment, installs all Python dependencies, and downloads
useful resources, including both sets of model parameters. Run:
scripts/install_third_party_dependencies.shTo activate the environment, run:
source scripts/activate_conda_env.shWith the environment active, compile CUDA kernels with
python3 setup.py installTo install the HH-suite to /usr/bin, run
scripts/install_hh_suite.shTo run feature generation pipeline from .fasta to feature.pkl on DeepMind's MSA and template database, run e.g.:
python ./scripts/extract_pkl_from_fas.py ./example_data/fasta/ ./example_data/features/where example_data is the directory containing example fasta . If jackhmmer,
hhblits, hhsearch and kalign are available at the default path of
/usr/bin, their binary_path command-line arguments can be dropped.
If you've already computed alignments for the query, you have the option to
skip the expensive alignment computation here with
--use_precomputed_alignments.
See example bash scripts in example_data/scripts
To run unit tests, use
scripts/run_unit_tests.shThe script is a thin wrapper around Python's unittest suite, and recognizes
unittest arguments. E.g., to run a specific test verbosely:
scripts/run_unit_tests.sh -v tests.test_modelCertain tests require that AlphaFold (v2.0.1) be installed in the same Python
environment. These run components of AlphaFold and OpenFold side by side and
ensure that output activations are adequately similar. For most modules, we
target a maximum pointwise difference of 1e-4.
If you find our open-sourced code & models helpful to your research, please also consider star🌟 and cite📑 this repo. Thank you for your support!
@misc{OpenComplex_code,
author={Jingcheng, Yu and Zhaoming, Chen and Zhaoqun, Li and Mingliang, Zeng and Wenjun, Lin and He, Huang and Qiwei, Ye},
title={Code of OpenComplex},
year={2022},
howpublished = {\url{https://github.com/baaihealth/OpenComplex}}
}
It is recommended to also cite OpenFold and AlphaFold.
Copyright 2022 BAAI.
Extended from AlphaFold and OpenFold, OpenComplex is licensed under the permissive Apache Licence, Version 2.0.
If you encounter problems using OpenComplex, feel free to create an issue! We also welcome pull requests from the community.
For help or issues using the repos, please submit a GitHub issue.
For other communications, please contact Qiwei Ye ([email protected]).