A tensorflow implementation of densenet with FCN for liver segmentation.
All the basic configurations of model and training process are listed in config.py and managed by EasyDict. We define our config parameters as __C which is defined in config.py.
Note: All the configs maybe reset in an extra file ./config/dice_skipv2.yml and it will cover the configs in config.py. We recommend to write a new .yml file for your own configuration instead of modifying config.py.
-
Dataset root directory is set as
__C.DATA.ROOT_DIR. You can specify root path both in windows or linux and the code will identify automatically. -
Then
__C.DATA.TRAINSET,__C.DATA.TESTSETand__C.DATA.VALSETshould be set as related directories of dataset. Here an example of directory tree is showed below.liverdirectory save all the 2D liver slices whilemaskdirectory save all the 2D mask slices.
./data
├─Liver_2017_train
│ ├─liver
│ └─mask
├─Liver_2017_val
│ ├─liver
│ └─mask
├─Liver_2017_test
│ ├─liver
│ └─mask
├─Liver_2017_test_3D
│ ├─liver
│ └─mask
-
All the liver and mask data is
.mhd+.rawformat. -
Set correct window width and level in
__C.IMG.W_WIDTHand__C.IMG.W_LEVEL.
-
Please set
__C.TAGa specify string to identify current model. -
If
__C.PRED_TAGis not empty, then the segmentation results will be stored. -
__C.TRAIN.MAX_ITERS: Total steps -
__C.MODELcontains all theFC-Densenetparameters such as growth rate, compression rate. -
__C.BACKBONE: We also implement U-Net and Tiramisu which can also be used as backbone. You can add your own network by inheritclass Networkwhich is defined innetworks.py. -
Other parameters please reference the description in
config.py.
Run the command below to train your model. This code will save the best weights(for validation) during training and the lastest weights. And dice_skipv2 means the specific config file defined in ./config/. 0 is the GPU ID you want to use. Only single GPU is supported.
bash main.sh train 0 dice_skipv2Run the command below to test your model in 2D mode with the best model.
bash main.sh test 0 dice_skipv2 2D trueRun the command below to test your model in 3D mode with the lastest model. 3D mode means that run 2D slices and concat them to 3D.
bash main.sh test 0 dice_skipv2 3D falseWe implement dice metric and other 5 metrics for evaluating segmentation accuracy. Reference
-
Dice coefficient
-
VOE
-
VD
-
ASD
-
RMSD
-
MSD
Example of public dataset 3D-Ircadb 01. Left part is label, right part is prediction.
| Liver-heart weak boundary | Isolated region |
|---|---|
 |
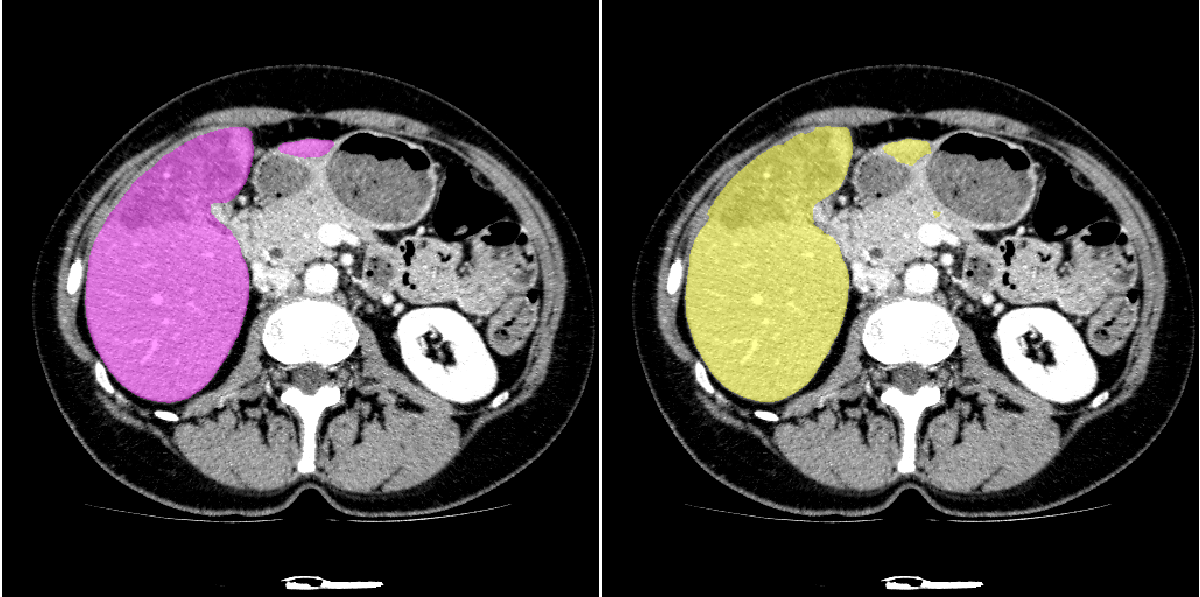 |
| Small region | Gap |
|---|---|
 |
 |
Maybe you want to freeze model with different batch size. You can set __C.TEST.SAVE_MODEL as true just like ./config/dice_skipv2_deploy.yml and change __C.TEST.BS_2D to other number. Then execute test routine:
bash main.sh test 0 dice_skipv2_deploy 2D trueit will save an empty model with new batch size. Then you will get three new file:
deploy_default_best.ckpt.data-00000-of-00001
deploy_default_best.ckpt.index
deploy_default_best.ckpt.meta
Of course we just need meta graph with new batch size deploy_default_best.ckpt.meta. Copy and replace the other two file with real weights:
cp default_best.ckpt.data-00000-of-00001 deploy_default_best.ckpt.data-00000-of-00001
cp default_best.ckpt.index deploy_default_best.ckpt.indexThen we can freeze model deploy_default_best.ckpt to deploy_default_best.pb. First change the first if to True in the __main__ part of ./utils/Model_Kits.py, then execute
CUDA_VISIBLE_DEVICES=0 python ./utils/Model_Kits.py --model dice_skipv2 --prefix deploy_defaultNote that --model argument denote the model tag __C.TAG while --prefix argument denote the model prefix __C.PREFIX. Re-generated model has a prefix deploy_xxxx.
Change the first if to False and the second if to True in the __main__ part of ./utils/Model_Kits.py, then execute
CUDA_VISIBLE_DEVICES=0 python ./utils/Model_Kits.pyFor details please read the segmentation() function in ./utils/Model_Kits.py.
Please make sure you have compile the tensorflow C++ API from source.
Create a new project demo and add load_graph.cpp to project, add include files, tensorflow.lib to VS configuration and add tensorflow.dll to environment variable PATH. Then compile and run
demo.exe path/to/xxx.pb xxx.raw1 xxx.raw2 ...