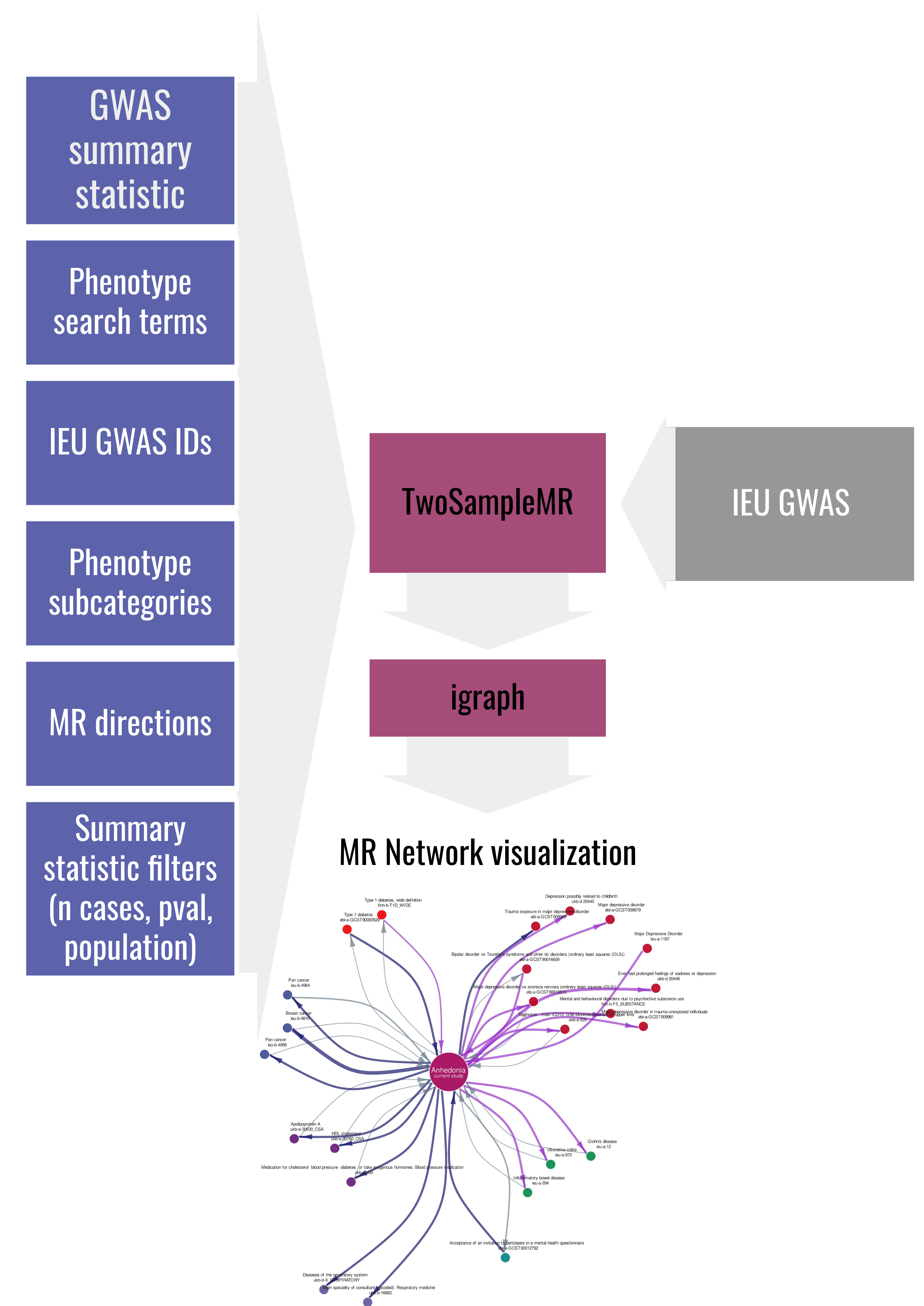
A script to run two-sample Mendelian Randomization (MR) in predefined directions using IEU GWAS data with provided in-house summary statistic in an automated manner.
- to run MR to assess the effect of the summary statistic of interest (./ex/GWAS_example.tsv) on a range of IEU phenotypes with count of cases>4000):
Rscript AutoMR.R -i './ex/GWAS_example.tsv' -r 'rsid' -e 'LOG_OR' -o 'OR' -s 'LOG(OR)_SE' -p 'P' --effect_allele_column 'ALT' --other_allele_column 'REF' -n 4520 -f TRUE -b FALSE --grep_phenotypes 'depress,bipol' --grep_subcategories 'Psychiatric / neurological' --specific_studies 'ieu-b-5075' -d 4000 --forward_pvals 5e-5 --visualize TRUE -x 'Anhedonia,current study'