Mpp6 Incorporation in the Nuclear Exosome Contributes to RNA Channeling through the Mtr4 Helicase.
Falk, S., Bonneau, F., Ebert, J., Kogel, A., Conti, E.(2017) Cell Rep 20: 2279-2286
- PubMed: 28877463
- DOI: https://doi.org/10.1016/j.celrep.2017.08.033
- Primary Citation of Related Structures:
5OKZ - PubMed Abstract:
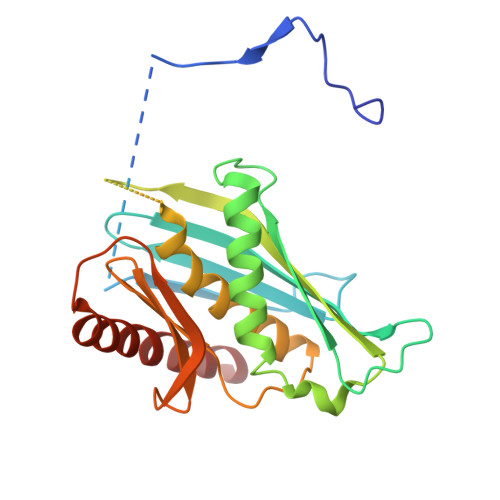
The RNA-degrading exosome mediates the processing and decay of many cellular transcripts. In the yeast nucleus, the ubiquitous 10-subunit exosome core complex (Exo-9-Rrp44) functions with four conserved cofactors (Rrp6, Rrp47, Mtr4, and Mpp6). Biochemical and structural studies to date have shed insights into the mechanisms of the exosome core and its nuclear cofactors, with the exception of Mpp6. We report the 3.2-Å resolution crystal structure of a S. cerevisiae Exo-9-Mpp6 complex, revealing how linear motifs in the Mpp6 middle domain bind Rrp40 via evolutionary conserved residues. In particular, Mpp6 binds near a tryptophan residue of Rrp40 that is mutated in human patients suffering from pontocerebellar hypoplasia. Using biochemical assays, we show that Mpp6 is required for the ability of Mtr4 to extend the trajectory of an RNA entering the exosome core, suggesting that it promotes the channeling of substrates from the nuclear helicase to the processive RNase.
Organizational Affiliation:
Max-Planck-Institute of Biochemistry, Department of Structural Cell Biology, Am Klopferspitz 18, 82152 Martinsried/Munich, Germany.