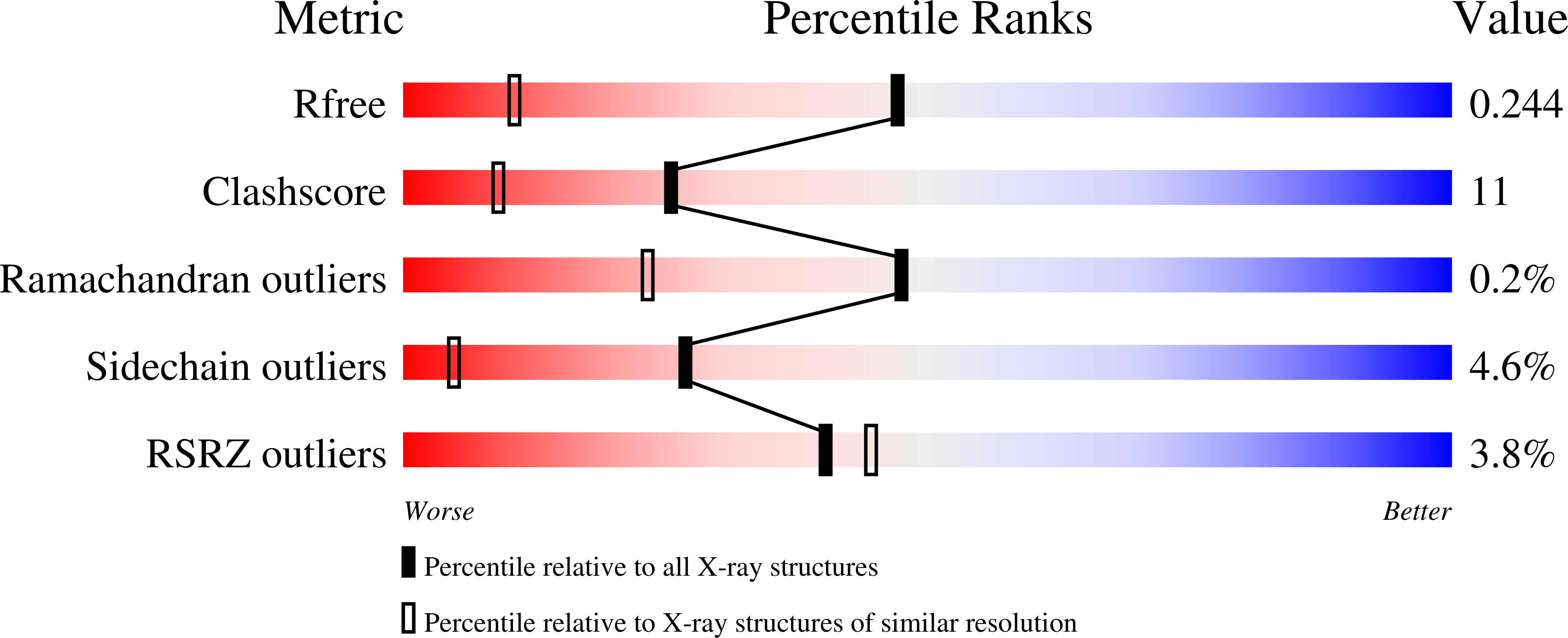
Structure of the lamin A/C R482W mutant responsible for dominant familial partial lipodystrophy (FPLD).
Magracheva, E., Kozlov, S., Stewart, C.L., Wlodawer, A., Zdanov, A.(2009) Acta Crystallogr Sect F Struct Biol Cryst Commun 65: 665-670
- PubMed: 19574635
- DOI: https://doi.org/10.1107/S1744309109020302
- Primary Citation of Related Structures:
3GEF - PubMed Abstract:
Proteins of the A-type lamin family, which consists of two members, lamin A and lamin C, are the major components of a thin proteinaceous filamentous meshwork, the lamina, that underlies the inner nuclear membrane. A-type lamins have recently become the focus of extensive functional studies as a consequence of the linking of at least eight congenital diseases to mutations in the lamin A/C gene (LMNA). This spectrum of pathologies, which mostly manifest themselves as dominant traits, includes muscle dystrophies, dilated cardiomyopathies, the premature aging syndrome Hutchinson-Guilford progeria and familial partial lipodystrophy (FPLD). The crystal structure of the lamin A/C mutant R482W, a variant that causes FPLD, has been determined at 1.5 A resolution. A completely novel aggregation state of the C-terminal globular domain and the position of the mutated amino-acid residue suggest means by which the mutation may affect lamin A/C-protein and protein-DNA interactions.
Organizational Affiliation:
Basic Research Program SAIC-Frederick, NCI-Frederick, Frederick, MD 21702, USA.