Structure of basic winged-bean lectin and a comparison with its saccharide-bound form.
Manoj, N., Srinivas, V.R., Suguna, K.(1999) Acta Crystallogr D Biol Crystallogr 55: 794-800
- PubMed: 10089310
- DOI: https://doi.org/10.1107/s090744499900044x
- Primary Citation of Related Structures:
1WBF - PubMed Abstract:
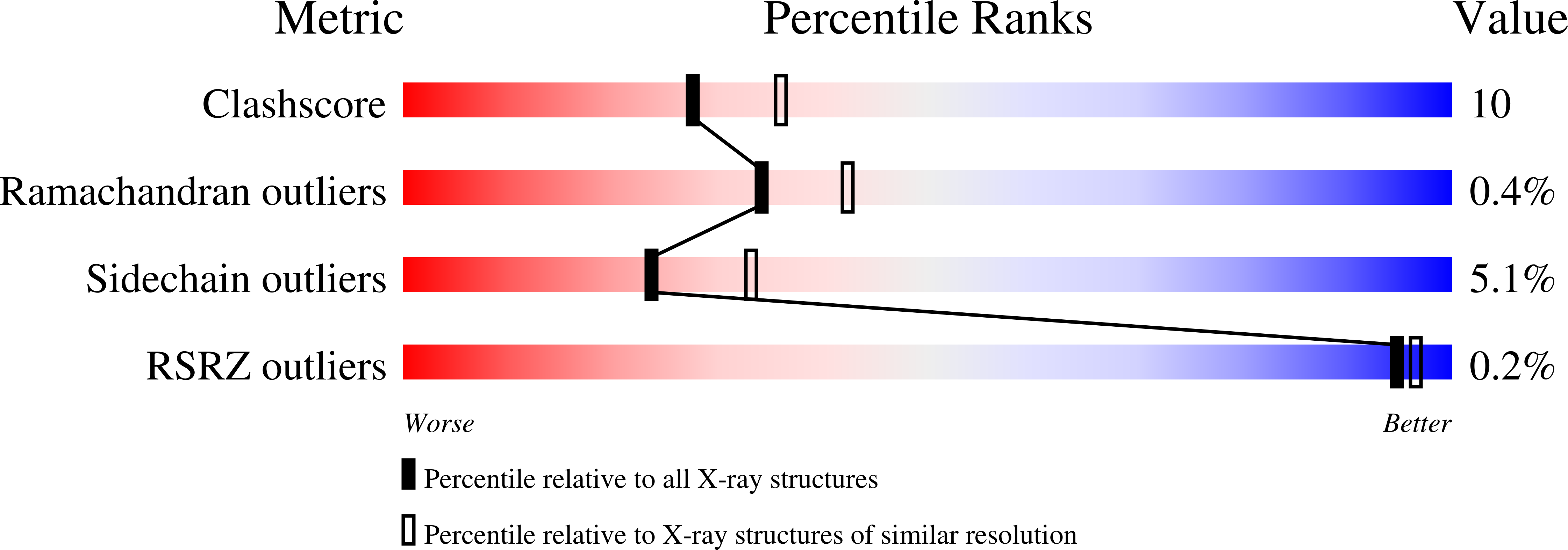
The crystal structure of the saccharide-free form of the basic form of winged-bean agglutinin (WBAI) has been solved by the molecular-replacement method and refined at 2.3 A resolution. The final R factor is 19.7% for all data in the resolution range 8.0-2.3 A. The asymmetric unit contains two half-dimers, each located on a crystallographic twofold axis. The structure of the saccharide-free form is compared with that of the complex of WBAI with methyl-alpha-D-galactoside. The complex is composed of two dimers in the asymmetric unit. The intersubunit interactions in the dimer are nearly identical in the two structures. The binding site of the saccharide-free structure contains three ordered water molecules at positions similar to those of the hydroxyl groups of the carbohydrate which are hydrogen bonded to the protein. Superposition of the saccharide-binding sites of the two structures shows that the major changes involve expulsion of these ordered water molecules and a shift of about 0.6 A of the main-chain atoms of the variable loop.
Organizational Affiliation:
Molecular Biophysics Unit, Indian Institute of Science, Bangalore-560 012, India.