WO2022227225A1 - 一种嵌合肽修饰的sis膜、其制备方法及应用 - Google Patents
一种嵌合肽修饰的sis膜、其制备方法及应用 Download PDFInfo
- Publication number
- WO2022227225A1 WO2022227225A1 PCT/CN2021/097851 CN2021097851W WO2022227225A1 WO 2022227225 A1 WO2022227225 A1 WO 2022227225A1 CN 2021097851 W CN2021097851 W CN 2021097851W WO 2022227225 A1 WO2022227225 A1 WO 2022227225A1
- Authority
- WO
- WIPO (PCT)
- Prior art keywords
- chimeric peptide
- sis
- seq
- membrane
- sequence shown
- Prior art date
Links
- 239000012528 membrane Substances 0.000 title claims abstract description 136
- 238000002360 preparation method Methods 0.000 title claims abstract description 21
- 230000007547 defect Effects 0.000 claims abstract description 34
- 210000000988 bone and bone Anatomy 0.000 claims abstract description 28
- 230000002188 osteogenic effect Effects 0.000 claims abstract description 22
- 230000000844 anti-bacterial effect Effects 0.000 claims abstract description 19
- 238000011282 treatment Methods 0.000 claims abstract description 13
- 208000015181 infectious disease Diseases 0.000 claims abstract description 10
- 230000002458 infectious effect Effects 0.000 claims abstract description 7
- 108091006116 chimeric peptides Proteins 0.000 claims description 93
- 238000000034 method Methods 0.000 claims description 37
- 239000000463 material Substances 0.000 claims description 34
- 230000000968 intestinal effect Effects 0.000 claims description 24
- 210000004876 tela submucosa Anatomy 0.000 claims description 24
- 230000035876 healing Effects 0.000 claims description 21
- 238000001035 drying Methods 0.000 claims description 11
- 239000012620 biological material Substances 0.000 claims description 9
- 230000002163 immunogen Effects 0.000 claims description 7
- 238000011221 initial treatment Methods 0.000 claims description 7
- 230000004048 modification Effects 0.000 claims description 6
- 238000012986 modification Methods 0.000 claims description 6
- 238000010030 laminating Methods 0.000 claims description 5
- 239000002904 solvent Substances 0.000 claims description 4
- 238000004519 manufacturing process Methods 0.000 claims description 2
- 230000014509 gene expression Effects 0.000 abstract description 20
- 108010024682 Core Binding Factor Alpha 1 Subunit Proteins 0.000 abstract description 8
- 102000015775 Core Binding Factor Alpha 1 Subunit Human genes 0.000 abstract description 8
- 241000194023 Streptococcus sanguinis Species 0.000 abstract description 7
- 241000194026 Streptococcus gordonii Species 0.000 abstract description 3
- 230000012010 growth Effects 0.000 abstract description 3
- 230000008827 biological function Effects 0.000 abstract description 2
- 101710189683 Alkaline protease 1 Proteins 0.000 abstract 1
- 101710154562 Alkaline proteinase Proteins 0.000 abstract 1
- 102100021253 Antileukoproteinase Human genes 0.000 abstract 1
- 101710170876 Antileukoproteinase Proteins 0.000 abstract 1
- 102100024506 Bone morphogenetic protein 2 Human genes 0.000 abstract 1
- 101710112538 C-C motif chemokine 27 Proteins 0.000 abstract 1
- 101000762366 Homo sapiens Bone morphogenetic protein 2 Proteins 0.000 abstract 1
- 101000613820 Homo sapiens Osteopontin Proteins 0.000 abstract 1
- 102100040557 Osteopontin Human genes 0.000 abstract 1
- 108090000765 processed proteins & peptides Proteins 0.000 description 47
- 239000000243 solution Substances 0.000 description 35
- 102000004196 processed proteins & peptides Human genes 0.000 description 33
- 108090000623 proteins and genes Proteins 0.000 description 32
- 238000006467 substitution reaction Methods 0.000 description 29
- 150000001413 amino acids Chemical class 0.000 description 28
- 229920001184 polypeptide Polymers 0.000 description 27
- 239000002773 nucleotide Substances 0.000 description 25
- 125000003729 nucleotide group Chemical group 0.000 description 25
- 108010035532 Collagen Proteins 0.000 description 24
- 102000008186 Collagen Human genes 0.000 description 24
- 229920001436 collagen Polymers 0.000 description 24
- 230000011164 ossification Effects 0.000 description 21
- 108091033319 polynucleotide Proteins 0.000 description 20
- 102000040430 polynucleotide Human genes 0.000 description 20
- 239000002157 polynucleotide Substances 0.000 description 20
- 238000004458 analytical method Methods 0.000 description 18
- 238000013508 migration Methods 0.000 description 17
- 230000035772 mutation Effects 0.000 description 17
- 101100244913 Mus musculus Prdm9 gene Proteins 0.000 description 15
- 102000004169 proteins and genes Human genes 0.000 description 15
- 230000005012 migration Effects 0.000 description 14
- 210000004027 cell Anatomy 0.000 description 13
- 238000002474 experimental method Methods 0.000 description 13
- 230000012292 cell migration Effects 0.000 description 12
- 108010009565 Bio-Gide Proteins 0.000 description 11
- 239000002609 medium Substances 0.000 description 11
- 238000010186 staining Methods 0.000 description 11
- 230000000694 effects Effects 0.000 description 10
- 241000894006 Bacteria Species 0.000 description 9
- 238000011529 RT qPCR Methods 0.000 description 9
- 230000010478 bone regeneration Effects 0.000 description 8
- 230000029663 wound healing Effects 0.000 description 8
- 102000002260 Alkaline Phosphatase Human genes 0.000 description 7
- 108020004774 Alkaline Phosphatase Proteins 0.000 description 7
- 102000008143 Bone Morphogenetic Protein 2 Human genes 0.000 description 7
- 108010049931 Bone Morphogenetic Protein 2 Proteins 0.000 description 7
- 102000004264 Osteopontin Human genes 0.000 description 7
- 108010081689 Osteopontin Proteins 0.000 description 7
- 241000700159 Rattus Species 0.000 description 7
- 125000000539 amino acid group Chemical group 0.000 description 7
- 238000003119 immunoblot Methods 0.000 description 7
- 239000000203 mixture Substances 0.000 description 7
- 238000004626 scanning electron microscopy Methods 0.000 description 7
- 210000004872 soft tissue Anatomy 0.000 description 7
- 102000012422 Collagen Type I Human genes 0.000 description 6
- 108010022452 Collagen Type I Proteins 0.000 description 6
- 206010052428 Wound Diseases 0.000 description 6
- 208000027418 Wounds and injury Diseases 0.000 description 6
- 230000005764 inhibitory process Effects 0.000 description 6
- 238000004445 quantitative analysis Methods 0.000 description 6
- 102000044503 Antimicrobial Peptides Human genes 0.000 description 5
- 108700042778 Antimicrobial Peptides Proteins 0.000 description 5
- DHMQDGOQFOQNFH-UHFFFAOYSA-N Glycine Chemical compound NCC(O)=O DHMQDGOQFOQNFH-UHFFFAOYSA-N 0.000 description 5
- 206010061218 Inflammation Diseases 0.000 description 5
- 102000004067 Osteocalcin Human genes 0.000 description 5
- 108090000573 Osteocalcin Proteins 0.000 description 5
- 108010087230 Sincalide Proteins 0.000 description 5
- 238000010609 cell counting kit-8 assay Methods 0.000 description 5
- 229940096422 collagen type i Drugs 0.000 description 5
- 238000013461 design Methods 0.000 description 5
- 238000010586 diagram Methods 0.000 description 5
- 238000003364 immunohistochemistry Methods 0.000 description 5
- 238000001727 in vivo Methods 0.000 description 5
- 230000004054 inflammatory process Effects 0.000 description 5
- 230000001105 regulatory effect Effects 0.000 description 5
- IZTQOLKUZKXIRV-YRVFCXMDSA-N sincalide Chemical compound C([C@@H](C(=O)N[C@@H](CCSC)C(=O)NCC(=O)N[C@@H](CC=1C2=CC=CC=C2NC=1)C(=O)N[C@@H](CCSC)C(=O)N[C@@H](CC(O)=O)C(=O)N[C@@H](CC=1C=CC=CC=1)C(N)=O)NC(=O)[C@@H](N)CC(O)=O)C1=CC=C(OS(O)(=O)=O)C=C1 IZTQOLKUZKXIRV-YRVFCXMDSA-N 0.000 description 5
- 238000001356 surgical procedure Methods 0.000 description 5
- 210000001519 tissue Anatomy 0.000 description 5
- 238000001262 western blot Methods 0.000 description 5
- 102000010834 Extracellular Matrix Proteins Human genes 0.000 description 4
- 108010037362 Extracellular Matrix Proteins Proteins 0.000 description 4
- ZHNUHDYFZUAESO-UHFFFAOYSA-N Formamide Chemical compound NC=O ZHNUHDYFZUAESO-UHFFFAOYSA-N 0.000 description 4
- 238000003559 RNA-seq method Methods 0.000 description 4
- 230000001580 bacterial effect Effects 0.000 description 4
- 210000000170 cell membrane Anatomy 0.000 description 4
- 238000012217 deletion Methods 0.000 description 4
- 230000037430 deletion Effects 0.000 description 4
- 238000010790 dilution Methods 0.000 description 4
- 239000012895 dilution Substances 0.000 description 4
- 210000002919 epithelial cell Anatomy 0.000 description 4
- 210000002744 extracellular matrix Anatomy 0.000 description 4
- 239000012091 fetal bovine serum Substances 0.000 description 4
- 239000000835 fiber Substances 0.000 description 4
- 230000006870 function Effects 0.000 description 4
- 238000007490 hematoxylin and eosin (H&E) staining Methods 0.000 description 4
- 238000010185 immunofluorescence analysis Methods 0.000 description 4
- 238000002991 immunohistochemical analysis Methods 0.000 description 4
- 239000007943 implant Substances 0.000 description 4
- 230000004807 localization Effects 0.000 description 4
- 210000000963 osteoblast Anatomy 0.000 description 4
- 230000008569 process Effects 0.000 description 4
- 230000001737 promoting effect Effects 0.000 description 4
- 239000000523 sample Substances 0.000 description 4
- 108020004414 DNA Proteins 0.000 description 3
- WSFSSNUMVMOOMR-UHFFFAOYSA-N Formaldehyde Chemical compound O=C WSFSSNUMVMOOMR-UHFFFAOYSA-N 0.000 description 3
- 102000003688 G-Protein-Coupled Receptors Human genes 0.000 description 3
- 108090000045 G-Protein-Coupled Receptors Proteins 0.000 description 3
- 241001465754 Metazoa Species 0.000 description 3
- 108091028043 Nucleic acid sequence Proteins 0.000 description 3
- 239000002033 PVDF binder Substances 0.000 description 3
- 208000006389 Peri-Implantitis Diseases 0.000 description 3
- 230000015572 biosynthetic process Effects 0.000 description 3
- 238000004113 cell culture Methods 0.000 description 3
- 230000004663 cell proliferation Effects 0.000 description 3
- 230000000295 complement effect Effects 0.000 description 3
- 210000002808 connective tissue Anatomy 0.000 description 3
- 238000012258 culturing Methods 0.000 description 3
- 238000005516 engineering process Methods 0.000 description 3
- 238000011156 evaluation Methods 0.000 description 3
- MHMNJMPURVTYEJ-UHFFFAOYSA-N fluorescein-5-isothiocyanate Chemical compound O1C(=O)C2=CC(N=C=S)=CC=C2C21C1=CC=C(O)C=C1OC1=CC(O)=CC=C21 MHMNJMPURVTYEJ-UHFFFAOYSA-N 0.000 description 3
- 239000012634 fragment Substances 0.000 description 3
- 238000000338 in vitro Methods 0.000 description 3
- 238000011534 incubation Methods 0.000 description 3
- 230000006698 induction Effects 0.000 description 3
- 230000008595 infiltration Effects 0.000 description 3
- 238000001764 infiltration Methods 0.000 description 3
- 230000007246 mechanism Effects 0.000 description 3
- 102000039446 nucleic acids Human genes 0.000 description 3
- 108020004707 nucleic acids Proteins 0.000 description 3
- 150000007523 nucleic acids Chemical class 0.000 description 3
- 201000001245 periodontitis Diseases 0.000 description 3
- 229920002981 polyvinylidene fluoride Polymers 0.000 description 3
- 230000035755 proliferation Effects 0.000 description 3
- PYWVYCXTNDRMGF-UHFFFAOYSA-N rhodamine B Chemical compound [Cl-].C=12C=CC(=[N+](CC)CC)C=C2OC2=CC(N(CC)CC)=CC=C2C=1C1=CC=CC=C1C(O)=O PYWVYCXTNDRMGF-UHFFFAOYSA-N 0.000 description 3
- 229940043267 rhodamine b Drugs 0.000 description 3
- 239000013598 vector Substances 0.000 description 3
- 238000005406 washing Methods 0.000 description 3
- HKZAAJSTFUZYTO-LURJTMIESA-N (2s)-2-[[2-[[2-[[2-[(2-aminoacetyl)amino]acetyl]amino]acetyl]amino]acetyl]amino]-3-hydroxypropanoic acid Chemical compound NCC(=O)NCC(=O)NCC(=O)NCC(=O)N[C@@H](CO)C(O)=O HKZAAJSTFUZYTO-LURJTMIESA-N 0.000 description 2
- JLDSMZIBHYTPPR-UHFFFAOYSA-N Alexa Fluor 405 Chemical compound CC[NH+](CC)CC.CC[NH+](CC)CC.CC[NH+](CC)CC.C12=C3C=4C=CC2=C(S([O-])(=O)=O)C=C(S([O-])(=O)=O)C1=CC=C3C(S(=O)(=O)[O-])=CC=4OCC(=O)N(CC1)CCC1C(=O)ON1C(=O)CCC1=O JLDSMZIBHYTPPR-UHFFFAOYSA-N 0.000 description 2
- 208000002679 Alveolar Bone Loss Diseases 0.000 description 2
- CIWBSHSKHKDKBQ-JLAZNSOCSA-N Ascorbic acid Chemical compound OC[C@H](O)[C@H]1OC(=O)C(O)=C1O CIWBSHSKHKDKBQ-JLAZNSOCSA-N 0.000 description 2
- 241000972773 Aulopiformes Species 0.000 description 2
- 108091003079 Bovine Serum Albumin Proteins 0.000 description 2
- 108010075254 C-Peptide Proteins 0.000 description 2
- 239000006144 Dulbecco’s modified Eagle's medium Substances 0.000 description 2
- KCXVZYZYPLLWCC-UHFFFAOYSA-N EDTA Chemical compound OC(=O)CN(CC(O)=O)CCN(CC(O)=O)CC(O)=O KCXVZYZYPLLWCC-UHFFFAOYSA-N 0.000 description 2
- 102000004190 Enzymes Human genes 0.000 description 2
- 108090000790 Enzymes Proteins 0.000 description 2
- DHCLVCXQIBBOPH-UHFFFAOYSA-N Glycerol 2-phosphate Chemical compound OCC(CO)OP(O)(O)=O DHCLVCXQIBBOPH-UHFFFAOYSA-N 0.000 description 2
- 108010072255 Integrin alpha3beta1 Proteins 0.000 description 2
- PIWKPBJCKXDKJR-UHFFFAOYSA-N Isoflurane Chemical compound FC(F)OC(Cl)C(F)(F)F PIWKPBJCKXDKJR-UHFFFAOYSA-N 0.000 description 2
- 229930040373 Paraformaldehyde Natural products 0.000 description 2
- KFSLWBXXFJQRDL-UHFFFAOYSA-N Peracetic acid Chemical compound CC(=O)OO KFSLWBXXFJQRDL-UHFFFAOYSA-N 0.000 description 2
- FAPWRFPIFSIZLT-UHFFFAOYSA-M Sodium chloride Chemical compound [Na+].[Cl-] FAPWRFPIFSIZLT-UHFFFAOYSA-M 0.000 description 2
- 238000002105 Southern blotting Methods 0.000 description 2
- 241000194017 Streptococcus Species 0.000 description 2
- 229920004890 Triton X-100 Polymers 0.000 description 2
- 239000013504 Triton X-100 Substances 0.000 description 2
- 241000700605 Viruses Species 0.000 description 2
- 230000033115 angiogenesis Effects 0.000 description 2
- 230000003110 anti-inflammatory effect Effects 0.000 description 2
- 230000009286 beneficial effect Effects 0.000 description 2
- 230000008901 benefit Effects 0.000 description 2
- 238000010170 biological method Methods 0.000 description 2
- 230000033558 biomineral tissue development Effects 0.000 description 2
- 239000012876 carrier material Substances 0.000 description 2
- 239000002299 complementary DNA Substances 0.000 description 2
- 210000000805 cytoplasm Anatomy 0.000 description 2
- 239000004053 dental implant Substances 0.000 description 2
- 238000000151 deposition Methods 0.000 description 2
- 230000008021 deposition Effects 0.000 description 2
- 239000002158 endotoxin Substances 0.000 description 2
- 210000002950 fibroblast Anatomy 0.000 description 2
- 238000004108 freeze drying Methods 0.000 description 2
- 238000003125 immunofluorescent labeling Methods 0.000 description 2
- 230000002757 inflammatory effect Effects 0.000 description 2
- 230000002401 inhibitory effect Effects 0.000 description 2
- 239000002054 inoculum Substances 0.000 description 2
- 238000003780 insertion Methods 0.000 description 2
- 230000037431 insertion Effects 0.000 description 2
- 229960002725 isoflurane Drugs 0.000 description 2
- 210000002510 keratinocyte Anatomy 0.000 description 2
- 229920006008 lipopolysaccharide Polymers 0.000 description 2
- 210000004698 lymphocyte Anatomy 0.000 description 2
- 238000010603 microCT Methods 0.000 description 2
- 230000000813 microbial effect Effects 0.000 description 2
- 238000010232 migration assay Methods 0.000 description 2
- 229920002866 paraformaldehyde Polymers 0.000 description 2
- 239000002245 particle Substances 0.000 description 2
- 229920000642 polymer Polymers 0.000 description 2
- 230000002062 proliferating effect Effects 0.000 description 2
- 230000026447 protein localization Effects 0.000 description 2
- 230000002441 reversible effect Effects 0.000 description 2
- 235000019515 salmon Nutrition 0.000 description 2
- 210000003491 skin Anatomy 0.000 description 2
- 210000003625 skull Anatomy 0.000 description 2
- 239000011780 sodium chloride Substances 0.000 description 2
- 239000001488 sodium phosphate Substances 0.000 description 2
- 229910000162 sodium phosphate Inorganic materials 0.000 description 2
- 238000007619 statistical method Methods 0.000 description 2
- 238000003786 synthesis reaction Methods 0.000 description 2
- RYFMWSXOAZQYPI-UHFFFAOYSA-K trisodium phosphate Chemical compound [Na+].[Na+].[Na+].[O-]P([O-])([O-])=O RYFMWSXOAZQYPI-UHFFFAOYSA-K 0.000 description 2
- 125000003088 (fluoren-9-ylmethoxy)carbonyl group Chemical group 0.000 description 1
- 108091032973 (ribonucleotides)n+m Proteins 0.000 description 1
- QMOQBVOBWVNSNO-UHFFFAOYSA-N 2-[[2-[[2-[(2-azaniumylacetyl)amino]acetyl]amino]acetyl]amino]acetate Chemical compound NCC(=O)NCC(=O)NCC(=O)NCC(O)=O QMOQBVOBWVNSNO-UHFFFAOYSA-N 0.000 description 1
- FWBHETKCLVMNFS-UHFFFAOYSA-N 4',6-Diamino-2-phenylindol Chemical compound C1=CC(C(=N)N)=CC=C1C1=CC2=CC=C(C(N)=N)C=C2N1 FWBHETKCLVMNFS-UHFFFAOYSA-N 0.000 description 1
- FWMNVWWHGCHHJJ-SKKKGAJSSA-N 4-amino-1-[(2r)-6-amino-2-[[(2r)-2-[[(2r)-2-[[(2r)-2-amino-3-phenylpropanoyl]amino]-3-phenylpropanoyl]amino]-4-methylpentanoyl]amino]hexanoyl]piperidine-4-carboxylic acid Chemical compound C([C@H](C(=O)N[C@H](CC(C)C)C(=O)N[C@H](CCCCN)C(=O)N1CCC(N)(CC1)C(O)=O)NC(=O)[C@H](N)CC=1C=CC=CC=1)C1=CC=CC=C1 FWMNVWWHGCHHJJ-SKKKGAJSSA-N 0.000 description 1
- 229920001817 Agar Polymers 0.000 description 1
- 208000035143 Bacterial infection Diseases 0.000 description 1
- 102000001187 Collagen Type III Human genes 0.000 description 1
- 108010069502 Collagen Type III Proteins 0.000 description 1
- 229920000742 Cotton Polymers 0.000 description 1
- 102000004127 Cytokines Human genes 0.000 description 1
- 108090000695 Cytokines Proteins 0.000 description 1
- 208000002064 Dental Plaque Diseases 0.000 description 1
- 102000007665 Extracellular Signal-Regulated MAP Kinases Human genes 0.000 description 1
- 108010007457 Extracellular Signal-Regulated MAP Kinases Proteins 0.000 description 1
- 108091006027 G proteins Proteins 0.000 description 1
- 102000030782 GTP binding Human genes 0.000 description 1
- 108091000058 GTP-Binding Proteins 0.000 description 1
- 102100031181 Glyceraldehyde-3-phosphate dehydrogenase Human genes 0.000 description 1
- 239000004471 Glycine Substances 0.000 description 1
- 102000003886 Glycoproteins Human genes 0.000 description 1
- 108090000288 Glycoproteins Proteins 0.000 description 1
- 108010061414 Hepatocyte Nuclear Factor 1-beta Proteins 0.000 description 1
- 102100022123 Hepatocyte nuclear factor 1-beta Human genes 0.000 description 1
- 101000979342 Homo sapiens Nuclear factor NF-kappa-B p105 subunit Proteins 0.000 description 1
- 108090001005 Interleukin-6 Proteins 0.000 description 1
- 102000004890 Interleukin-8 Human genes 0.000 description 1
- 108090001007 Interleukin-8 Proteins 0.000 description 1
- 102000009664 Microtubule-Associated Proteins Human genes 0.000 description 1
- 108010020004 Microtubule-Associated Proteins Proteins 0.000 description 1
- 206010028116 Mucosal inflammation Diseases 0.000 description 1
- 201000010927 Mucositis Diseases 0.000 description 1
- 206010028980 Neoplasm Diseases 0.000 description 1
- 102100023050 Nuclear factor NF-kappa-B p105 subunit Human genes 0.000 description 1
- 206010048685 Oral infection Diseases 0.000 description 1
- 101710116435 Outer membrane protein Proteins 0.000 description 1
- 206010034133 Pathogen resistance Diseases 0.000 description 1
- 108091000080 Phosphotransferase Proteins 0.000 description 1
- 241000605862 Porphyromonas gingivalis Species 0.000 description 1
- 102000001253 Protein Kinase Human genes 0.000 description 1
- 102000007056 Recombinant Fusion Proteins Human genes 0.000 description 1
- 108010008281 Recombinant Fusion Proteins Proteins 0.000 description 1
- MTCFGRXMJLQNBG-UHFFFAOYSA-N Serine Natural products OCC(N)C(O)=O MTCFGRXMJLQNBG-UHFFFAOYSA-N 0.000 description 1
- 208000008312 Tooth Loss Diseases 0.000 description 1
- 238000002835 absorbance Methods 0.000 description 1
- 239000008272 agar Substances 0.000 description 1
- 239000012670 alkaline solution Substances 0.000 description 1
- 238000000540 analysis of variance Methods 0.000 description 1
- 230000002924 anti-infective effect Effects 0.000 description 1
- 229940124350 antibacterial drug Drugs 0.000 description 1
- 239000004599 antimicrobial Substances 0.000 description 1
- 238000013459 approach Methods 0.000 description 1
- 229940072107 ascorbate Drugs 0.000 description 1
- 235000010323 ascorbic acid Nutrition 0.000 description 1
- 239000011668 ascorbic acid Substances 0.000 description 1
- 238000003556 assay Methods 0.000 description 1
- 208000022362 bacterial infectious disease Diseases 0.000 description 1
- 230000004888 barrier function Effects 0.000 description 1
- 230000032770 biofilm formation Effects 0.000 description 1
- 229920001222 biopolymer Polymers 0.000 description 1
- 230000000903 blocking effect Effects 0.000 description 1
- 230000010072 bone remodeling Effects 0.000 description 1
- 229940098773 bovine serum albumin Drugs 0.000 description 1
- 210000004556 brain Anatomy 0.000 description 1
- 238000004364 calculation method Methods 0.000 description 1
- 125000002091 cationic group Chemical group 0.000 description 1
- 150000001768 cations Chemical class 0.000 description 1
- 230000021164 cell adhesion Effects 0.000 description 1
- 230000030833 cell death Effects 0.000 description 1
- 230000003833 cell viability Effects 0.000 description 1
- 108091092328 cellular RNA Proteins 0.000 description 1
- 238000005119 centrifugation Methods 0.000 description 1
- 239000003153 chemical reaction reagent Substances 0.000 description 1
- 238000010367 cloning Methods 0.000 description 1
- 238000007621 cluster analysis Methods 0.000 description 1
- 230000001332 colony forming effect Effects 0.000 description 1
- 239000013078 crystal Substances 0.000 description 1
- 230000001086 cytosolic effect Effects 0.000 description 1
- 230000034994 death Effects 0.000 description 1
- 230000007123 defense Effects 0.000 description 1
- 230000018044 dehydration Effects 0.000 description 1
- 238000006297 dehydration reaction Methods 0.000 description 1
- 238000001514 detection method Methods 0.000 description 1
- 239000003599 detergent Substances 0.000 description 1
- UREBDLICKHMUKA-CXSFZGCWSA-N dexamethasone Chemical compound C1CC2=CC(=O)C=C[C@]2(C)[C@]2(F)[C@@H]1[C@@H]1C[C@@H](C)[C@@](C(=O)CO)(O)[C@@]1(C)C[C@@H]2O UREBDLICKHMUKA-CXSFZGCWSA-N 0.000 description 1
- 229960003957 dexamethasone Drugs 0.000 description 1
- 230000007646 directional migration Effects 0.000 description 1
- 238000011978 dissolution method Methods 0.000 description 1
- 239000003814 drug Substances 0.000 description 1
- 229940079593 drug Drugs 0.000 description 1
- 238000001962 electrophoresis Methods 0.000 description 1
- 230000009881 electrostatic interaction Effects 0.000 description 1
- 210000002889 endothelial cell Anatomy 0.000 description 1
- 210000001339 epidermal cell Anatomy 0.000 description 1
- 230000010393 epithelial cell migration Effects 0.000 description 1
- UCZQDTXHYSNFGD-UHFFFAOYSA-N ethaneperoxoic acid;ethanol Chemical compound CCO.CC(=O)OO UCZQDTXHYSNFGD-UHFFFAOYSA-N 0.000 description 1
- 239000013604 expression vector Substances 0.000 description 1
- 239000007850 fluorescent dye Substances 0.000 description 1
- 238000001215 fluorescent labelling Methods 0.000 description 1
- 108020001507 fusion proteins Proteins 0.000 description 1
- 102000037865 fusion proteins Human genes 0.000 description 1
- 108020004445 glyceraldehyde-3-phosphate dehydrogenase Proteins 0.000 description 1
- 108010001064 glycyl-glycyl-glycyl-glycine Proteins 0.000 description 1
- PCHJSUWPFVWCPO-UHFFFAOYSA-N gold Chemical compound [Au] PCHJSUWPFVWCPO-UHFFFAOYSA-N 0.000 description 1
- 239000010931 gold Substances 0.000 description 1
- 229910052737 gold Inorganic materials 0.000 description 1
- 238000012165 high-throughput sequencing Methods 0.000 description 1
- 238000003384 imaging method Methods 0.000 description 1
- 230000005847 immunogenicity Effects 0.000 description 1
- 230000006872 improvement Effects 0.000 description 1
- 230000000415 inactivating effect Effects 0.000 description 1
- 230000028709 inflammatory response Effects 0.000 description 1
- 238000001802 infusion Methods 0.000 description 1
- 208000014674 injury Diseases 0.000 description 1
- 230000015788 innate immune response Effects 0.000 description 1
- 108010044426 integrins Proteins 0.000 description 1
- 102000006495 integrins Human genes 0.000 description 1
- 230000003993 interaction Effects 0.000 description 1
- 238000005342 ion exchange Methods 0.000 description 1
- 108010028309 kalinin Proteins 0.000 description 1
- 230000029774 keratinocyte migration Effects 0.000 description 1
- 239000002648 laminated material Substances 0.000 description 1
- 238000011068 loading method Methods 0.000 description 1
- 230000007774 longterm Effects 0.000 description 1
- 210000002901 mesenchymal stem cell Anatomy 0.000 description 1
- 108020004999 messenger RNA Proteins 0.000 description 1
- 239000003226 mitogen Substances 0.000 description 1
- 238000002715 modification method Methods 0.000 description 1
- 238000010369 molecular cloning Methods 0.000 description 1
- 230000000877 morphologic effect Effects 0.000 description 1
- 238000001543 one-way ANOVA Methods 0.000 description 1
- 230000010355 oscillation Effects 0.000 description 1
- 238000010883 osseointegration Methods 0.000 description 1
- 238000010422 painting Methods 0.000 description 1
- 206010033675 panniculitis Diseases 0.000 description 1
- 239000012188 paraffin wax Substances 0.000 description 1
- 230000001936 parietal effect Effects 0.000 description 1
- 230000037361 pathway Effects 0.000 description 1
- 238000010647 peptide synthesis reaction Methods 0.000 description 1
- 208000004480 periapical periodontitis Diseases 0.000 description 1
- 230000035699 permeability Effects 0.000 description 1
- 239000008363 phosphate buffer Substances 0.000 description 1
- 150000003904 phospholipids Chemical class 0.000 description 1
- 230000026731 phosphorylation Effects 0.000 description 1
- 238000006366 phosphorylation reaction Methods 0.000 description 1
- 102000020233 phosphotransferase Human genes 0.000 description 1
- 239000003910 polypeptide antibiotic agent Substances 0.000 description 1
- 239000011148 porous material Substances 0.000 description 1
- 230000002265 prevention Effects 0.000 description 1
- 238000012545 processing Methods 0.000 description 1
- 230000004853 protein function Effects 0.000 description 1
- 108060006633 protein kinase Proteins 0.000 description 1
- 238000003753 real-time PCR Methods 0.000 description 1
- 230000008929 regeneration Effects 0.000 description 1
- 238000011069 regeneration method Methods 0.000 description 1
- 238000007634 remodeling Methods 0.000 description 1
- 230000008439 repair process Effects 0.000 description 1
- 238000010839 reverse transcription Methods 0.000 description 1
- 210000003079 salivary gland Anatomy 0.000 description 1
- 239000012266 salt solution Substances 0.000 description 1
- 238000004621 scanning probe microscopy Methods 0.000 description 1
- 238000006748 scratching Methods 0.000 description 1
- 230000002393 scratching effect Effects 0.000 description 1
- 238000000926 separation method Methods 0.000 description 1
- 230000019491 signal transduction Effects 0.000 description 1
- 238000002791 soaking Methods 0.000 description 1
- 239000007790 solid phase Substances 0.000 description 1
- 238000007711 solidification Methods 0.000 description 1
- 230000008023 solidification Effects 0.000 description 1
- 241000894007 species Species 0.000 description 1
- 238000012453 sprague-dawley rat model Methods 0.000 description 1
- 238000005507 spraying Methods 0.000 description 1
- 238000004544 sputter deposition Methods 0.000 description 1
- 210000004304 subcutaneous tissue Anatomy 0.000 description 1
- 239000000126 substance Substances 0.000 description 1
- 239000000758 substrate Substances 0.000 description 1
- 238000012360 testing method Methods 0.000 description 1
- 230000008467 tissue growth Effects 0.000 description 1
- 230000017423 tissue regeneration Effects 0.000 description 1
- 238000012546 transfer Methods 0.000 description 1
- 230000009466 transformation Effects 0.000 description 1
- 230000007704 transition Effects 0.000 description 1
- 230000008733 trauma Effects 0.000 description 1
- 238000002604 ultrasonography Methods 0.000 description 1
- 238000012800 visualization Methods 0.000 description 1
- XLYOFNOQVPJJNP-UHFFFAOYSA-N water Substances O XLYOFNOQVPJJNP-UHFFFAOYSA-N 0.000 description 1
- 230000010388 wound contraction Effects 0.000 description 1
- 230000037314 wound repair Effects 0.000 description 1
Images
Classifications
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61L—METHODS OR APPARATUS FOR STERILISING MATERIALS OR OBJECTS IN GENERAL; DISINFECTION, STERILISATION OR DEODORISATION OF AIR; CHEMICAL ASPECTS OF BANDAGES, DRESSINGS, ABSORBENT PADS OR SURGICAL ARTICLES; MATERIALS FOR BANDAGES, DRESSINGS, ABSORBENT PADS OR SURGICAL ARTICLES
- A61L27/00—Materials for grafts or prostheses or for coating grafts or prostheses
- A61L27/14—Macromolecular materials
- A61L27/22—Polypeptides or derivatives thereof, e.g. degradation products
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61L—METHODS OR APPARATUS FOR STERILISING MATERIALS OR OBJECTS IN GENERAL; DISINFECTION, STERILISATION OR DEODORISATION OF AIR; CHEMICAL ASPECTS OF BANDAGES, DRESSINGS, ABSORBENT PADS OR SURGICAL ARTICLES; MATERIALS FOR BANDAGES, DRESSINGS, ABSORBENT PADS OR SURGICAL ARTICLES
- A61L27/00—Materials for grafts or prostheses or for coating grafts or prostheses
- A61L27/36—Materials for grafts or prostheses or for coating grafts or prostheses containing ingredients of undetermined constitution or reaction products thereof, e.g. transplant tissue, natural bone, extracellular matrix
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61L—METHODS OR APPARATUS FOR STERILISING MATERIALS OR OBJECTS IN GENERAL; DISINFECTION, STERILISATION OR DEODORISATION OF AIR; CHEMICAL ASPECTS OF BANDAGES, DRESSINGS, ABSORBENT PADS OR SURGICAL ARTICLES; MATERIALS FOR BANDAGES, DRESSINGS, ABSORBENT PADS OR SURGICAL ARTICLES
- A61L27/00—Materials for grafts or prostheses or for coating grafts or prostheses
- A61L27/50—Materials characterised by their function or physical properties, e.g. injectable or lubricating compositions, shape-memory materials, surface modified materials
- A61L27/54—Biologically active materials, e.g. therapeutic substances
-
- C—CHEMISTRY; METALLURGY
- C07—ORGANIC CHEMISTRY
- C07K—PEPTIDES
- C07K19/00—Hybrid peptides, i.e. peptides covalently bound to nucleic acids, or non-covalently bound protein-protein complexes
Definitions
- the present disclosure belongs to the field of biomedical materials, and in particular relates to a SIS membrane combined with a chimeric peptide, a preparation method and application thereof.
- GBR Guided Bone Regeneration
- Extracellular matrix (ECM) material is an acellular biological network composed of collagen (mainly type I and type III) and various glycoproteins, and has become a hot spot for tissue engineering scaffolds in recent years.
- Small intestinal submucosa (SIS) is the most commonly used ECM material due to its wide source and easy availability.
- SIS has many excellent properties, including site-specific tissue regeneration capacity, low immunogenicity, excellent mechanical properties, and the inclusion of multiple cytokines. It has been used as a scaffold material in many types of tissues with satisfactory results.
- infectious bone defects caused by periodontitis, peri-implantitis, and apical periodontitis are a problem for GBR.
- Despite the excellent mechanical properties and biocompatibility of SIS membranes, their antibacterial, osteogenic, and healing-promoting abilities are limited. To address these limitations, it is hoped to find a suitable way for the modification of SIS.
- HST Histamines
- JH8194 also has good antibacterial activity and can effectively prevent peri-implantitis and peri-implant mucositis.
- JH8194 also has a certain ability to promote bone formation. It has been found that JH8194 can stimulate the expression of Runx2, Opn and Alp in MC3T3-E1 while inhibiting the biofilm formation of Pg, so that the cells can differentiate into osteoblasts.
- JH8194 can increase the formation of mature trabecular bone around the implant and enhance osseointegration, which is beneficial to the long-term stability of the implant. Therefore, JH8194 is an effective drug for antibacterial and osteogenic modification of SIS membranes.
- Hst1 another member of the HST, Hst1, displayed an excellent ability to promote healing.
- Hst1 can promote the migration or adhesion of fibroblasts, epithelial cells and endothelial cells, as well as guide epidermal cell regeneration and angiogenesis in the wound area.
- the present disclosure provides a chimeric peptide-modified SIS membrane that can simultaneously exert antibacterial effects, promote soft tissue healing and bone regeneration, and greatly enrich the performance of the GBR membrane.
- Synthetic peptide-modified SIS membranes ie, GBR membranes
- GBR membranes can be used for clinical treatment of infectious bone defects.
- the present disclosure describes the following technical solutions.
- a SIS membrane modified with a chimeric peptide wherein the sequence of the chimeric peptide comprises at least one of the group consisting of the following sequences:
- the sequence of the chimeric peptide in (1) comprises and the sequence shown in SEQ ID NO:9, the sequence shown in SEQ ID NO:10, and the sequence shown in SEQ ID NO:11 Compared with the sequence shown in SEQ ID NO: 12, the sequence has at least 80% identity.
- the sequence of the chimeric peptide in (1) comprises the sequence shown in SEQ ID NO: 9, the sequence shown in SEQ ID NO: 10, and the sequence shown in SEQ ID NO: 11
- the sequence shown, compared to the sequence shown in SEQ ID NO: 12, has 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90% %, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% or more identical sequences.
- the sequence of the chimeric peptide in (1) comprises the sequence shown in SEQ ID NO: 9, the sequence shown in SEQ ID NO: 10, and the sequence shown in SEQ ID NO: Compared with the sequence shown in SEQ ID NO: 12, the sequence shown in 11 has 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, A sequence of 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98% or 99% identity; and, it and the sequence shown in SEQ ID NO:9 , compared with the sequence shown in SEQ ID NO: 10, the sequence shown in SEQ ID NO: 11, and the sequence shown in SEQ ID NO: 12, there are only conservative substitutions.
- sequence of the chimeric peptide in (2) and the sequence shown in SEQ ID NO: 9, the sequence shown in SEQ ID NO: 10, and the sequence shown in SEQ ID NO: 11 The sequence shown, compared to the sequence shown in SEQ ID NO: 12, has 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90% %, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98% or more than 99% identity; Compared with the sequence shown in ID NO: 10, the sequence shown in SEQ ID NO: 11, and the sequence shown in SEQ ID NO: 12, there are only conservative substitutions.
- step (b) applying the dissolving solution obtained in step (a) to the surface of the SIS membrane; preferably, immersing the SIS membrane in the dissolving solution obtained in step (a);
- step (ii) subjecting the small intestinal submucosa material obtained in step (i) to immunogen removal treatment.
- step (b) applying the dissolving solution obtained in step (a) to the surface of the SIS membrane; preferably, immersing the SIS membrane in the dissolving solution obtained in step (a);
- sequence of the chimeric peptide comprises at least one of the group consisting of the following sequences, or the sequence of the chimeric peptide consists of at least one of the group consisting of the following sequences:
- the sequence of the chimeric peptide in (6) comprises the sequence shown in SEQ ID NO: 9, the sequence shown in SEQ ID NO: 10, and the sequence shown in SEQ ID NO: 11 Compared with the sequence shown in SEQ ID NO: 12, the sequence has at least 80% identity; or, the sequence of the chimeric peptide in (6) and the sequence shown in SEQ ID NO: 9 Compared with the sequence shown in SEQ ID NO: 10, the sequence shown in SEQ ID NO: 11, and the sequence shown in SEQ ID NO: 12, they have at least 80% identity.
- the sequence of the chimeric peptide in (6) comprises the sequence shown in SEQ ID NO:9, the sequence shown in SEQ ID NO:10, and the sequence shown in SEQ ID NO:11
- the sequence shown, compared to the sequence shown in SEQ ID NO: 12, has 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90% %, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98% or more than 99% identical; or, the sequence of the chimeric peptide in (6) and Compared with the sequence shown in SEQ ID NO: 9, the sequence shown in SEQ ID NO: 10, the sequence shown in SEQ ID NO: 11, and the sequence shown in SEQ ID NO: 12, it has 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97% , 98% or more
- the sequence of the chimeric peptide in (6) comprises the sequence shown in SEQ ID NO: 9, the sequence shown in SEQ ID NO: 10, and the sequence shown in SEQ ID NO: Compared with the sequence shown in SEQ ID NO: 12, the sequence shown in 11 has 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, A sequence of 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98% or 99% identity; and, it and the sequence shown in SEQ ID NO:9 , compared with the sequence shown in SEQ ID NO: 10, the sequence shown in SEQ ID NO: 11, and the sequence shown in SEQ ID NO: 12, there are only conservative substitutions.
- sequence of the chimeric peptide in (6) and the sequence shown in SEQ ID NO: 9, the sequence shown in SEQ ID NO: 10, and the sequence shown in SEQ ID NO: 11 The sequence shown, compared to the sequence shown in SEQ ID NO: 12, has 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90% %, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98% or more than 99% identity; Compared with the sequence shown in ID NO: 10, the sequence shown in SEQ ID NO: 11, and the sequence shown in SEQ ID NO: 12, there are only conservative substitutions.
- step (ii) subjecting the small intestinal submucosa material obtained in step (i) to immunogen removal treatment.
- a method of treating an infectious bone defect comprising administering to a subject the chimeric peptide-modified SIS membrane according to any one of (1)-(5) or according to (6) -(8) The step of the chimeric peptide-modified SIS membrane obtained by the production method of the chimeric peptide-modified SIS membrane according to any one of the above.
- the present disclosure successfully develops SIS membranes modified with chimeric peptides for the preparation of chimeric peptide-modified GBR membranes.
- the present disclosure employs a set of antibacterial, osteogenic, and healing-promoting chimeric peptides to target the surface of SIS membranes via collagen-binding sequences.
- the present disclosure utilizes a chimeric peptide-modified SIS membrane that promotes the expression of ITG- ⁇ 3, ITG- ⁇ 1, BMP2, RUNX2, ALP, and OPN, and inhibits S. gasseri and S. sanguis the growth of SIS membranes, thereby enabling the SIS membrane to exert antibacterial, osteogenic and healing-promoting biological functions.
- the SIS membrane modified with a chimeric peptide ie, a chimeric peptide-modified GBR membrane
- a chimeric peptide ie, a chimeric peptide-modified GBR membrane
- the SIS membrane modified with a chimeric peptide can be used for clinical treatment of infectious bone defects.
- Figure 1A shows the design details and structures of the collagen chimeric peptides P9 and P10. Among them, the legends are folded strand (Strand), helix (Helix) and random coil (Coil). Conf stands for Confidence of prediction; Cart stands for 3-state assignment cartoon; Pred stands for 3-state prediction; AA stands for Target Sequence.
- Figure IB shows the design details and structures of the collagen chimeric peptides P11 and P12. Among them, the legends are folded strand (Strand), helix (Helix) and random coil (Coil). Conf stands for Confidence of prediction; Cart stands for 3-state assignment cartoon; Pred stands for 3-state prediction; AA stands for Target Sequence.
- Figure 2A shows a schematic diagram of a pSIS membrane.
- Figure 2B shows the binding of chimeric peptides observed by CLSM.
- Figure 2C shows the surface morphology of lyophilized SIS and pSIS observed by SEM.
- Figure 2D shows the proliferative capacity of BMSCs and OECs by CCK-8.
- Figure 3A shows the zone of inhibition of S. sanguis and S. birin.
- Figure 3B shows CLSM images stained after culturing mixed bacteria in blank wells, SIS, M-pSIS or H-pSIS for 24 hours.
- Figure 4A shows the results of qPCR analysis of osteogenesis-related genes (* indicates p ⁇ 0.05).
- Figure 4B shows the results of immunoblot analysis of osteogenesis-related proteins.
- Figure 4C shows the quantitative analysis results of immunoblot analysis (* indicates p ⁇ 0.05).
- Figure 4D shows the results of immunofluorescence analysis of the localization of osteogenesis-related proteins in BMSCs (BMP2, RUNX2, ALP, OPN and nucleus).
- Figure 5A shows the results of the Transwell assay to detect cell migration ability.
- Figure 5B shows the results of the analysis of migrated cells (* indicates p ⁇ 0.05).
- Figure 5C shows the results of differentially expressed genes screened by RNA-seq.
- Figure 5D shows a Venn diagram of different genes.
- Figure 5E shows the results of qPCR analysis of migration-related genes.
- Figure 5F shows the results of Western blot analysis of migration-related proteins.
- Figure 5G shows the results of quantitative analysis of Western blot analysis (* indicates p ⁇ 0.05).
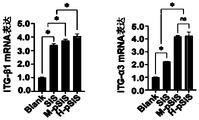
- Figure 5H shows the results of immunofluorescence analysis of migration-related protein localization in OECs (ITG-[alpha]3, ITG-[beta]1 and nucleus).
- Figure 6A shows three-dimensional reconstructions and sagittal images of bone defects covered with SIS, Bio-Gide and H-pSIS membranes.
- Figure 6B shows BV/TV results for different groups of bone defects (* indicates p ⁇ 0.05).
- FIG. 6C shows H&E staining results.
- Figure 6D shows the results of Masson's trichrome staining.
- Figure 6E shows the results of immunohistochemical analysis of OCN and COL1 expression.
- Figure 7A shows H&E staining results (line segment: length of non-epithelial wound).
- Figure 7B shows the length of neo-epithelialization (* indicates p ⁇ 0.05).
- Figure 7C shows the results of Masson's trichrome staining (line segment: length of non-collagenous fibers of the wound).
- Figure 7D shows collagen fiber length (* indicates p ⁇ 0.05).
- FIG. 7E shows the results of immunohistochemical analysis of ITG- ⁇ 3 and ITG- ⁇ 1 expression.
- Figure 8 shows the results of healing in rat experiments 1 and 2 weeks after surgery.
- Figure 9 shows a comparison of the osteogenic effects of SIS membranes modified with different chimeric peptides.
- Figure 10 shows a comparison of the anti-inflammatory effects of SIS membranes modified with different chimeric peptides.
- Figure 11 shows a comparison of the healing effects of SIS membranes modified with different chimeric peptides.
- amino acid mutation or “nucleotide mutation” includes “substitution, duplication, deletion or addition of one or more amino acids or nucleotides”.
- mutation refers to changes in nucleotide sequence or amino acid sequence.
- a “mutation” of the present disclosure may be selected from “conservative mutation”, “semi-conservative mutation”, “non-conservative mutation”.
- non-conservative mutation or “semi-conservative mutation” may be a mutation that results in a loss or partial loss of protein function.
- conservative mutation refers to a mutation that normally maintains the function of a protein. Representative examples of conservative mutations are conservative substitutions.
- substitutions typically exchange one amino acid at one or more sites in a protein. Such substitutions can be conservative. Specifically, substitutions regarded as conservative substitutions include substitution of Ala to Ser or Thr, substitution of Arg to Gln, His, or Lys, substitution of Asn to Glu, Gln, Lys, His, or Asp, substitution of Asp to Glu, Gin, Lys, His, or Asp.
- Sequence identity and “percent identity” in the present disclosure refer to the percentage of nucleotides or amino acids that are identical (ie, identical) between two or more polynucleotides or polypeptides. Sequence identity between two or more polynucleotides or polypeptides can be determined by aligning the nucleotide or amino acid sequences of the polynucleotides or polypeptides and The number of positions containing the same nucleotide or amino acid residue is scored and compared to the number of positions containing different nucleotide or amino acid residues in the aligned polynucleotides or polypeptides.
- Polynucleotides can differ at a position, for example, by containing different nucleotides (i.e., substitutions or mutations) or deletions of nucleotides (i.e., nucleotide insertions or nucleotide deletions in one or both polynucleotides).
- Polypeptides can differ at one position, for example, by containing different amino acids (ie, substitutions or mutations) or missing amino acids (ie, amino acid insertions or amino acid deletions in one or both polypeptides).
- Sequence identity can be calculated by dividing the number of positions containing the same nucleotide or amino acid residue by the total number of amino acid residues in a polynucleotide or polypeptide. For example, percent identity can be calculated by dividing the number of positions containing the same nucleotide or amino acid residue by the total number of nucleotides or amino acid residues in the polynucleotide or polypeptide and multiplying by 100.
- two or more sequences or subsequences have at least 40%, 50%, 60%, 50%, 60%, 40%, 60%, 60%, 50%, 60% %, 70%, 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% of the "sequence" of nucleotides or amino acid residues Identity" or "Percent Identity”.
- the determination/calculation of "sequence identity” or “percent identity” can be based on any suitable region of the sequence.
- sequences are substantially identical over the entire length of either or both of the compared biopolymers (ie, nucleic acids or polypeptides).
- Reverse Complementary Sequence means a sequence that is oriented in the opposite direction to the sequence of the original polynucleotide and is also complementary to the sequence of the original polynucleotide. Exemplarily, if the original polynucleotide sequence is ACTGAAC, its reverse complement is GTTCAGT.
- polynucleotide refers to a polymer composed of nucleotides.
- Polynucleotides may be in the form of individual fragments or part of a larger nucleotide sequence structure derived from a nucleotide sequence that has been isolated at least once in quantity or concentration, and can be obtained by standard Molecular biological methods (eg, using cloning vectors) identify, manipulate, and recover sequences and their component nucleotide sequences.
- a nucleotide sequence is represented by a DNA sequence (ie A, T, G, C)
- this also includes an RNA sequence (ie A, U, G, C) where "U" replaces "T”.
- polynucleotide refers to a polymer of nucleotides removed from other nucleotides (individual fragments or entire fragments), or may be a component or component of a larger nucleotide structure, such as an expression vector or polycistronic sequence.
- Polynucleotides include DNA, RNA and cDNA sequences.
- Recombinant polynucleotide and “recombinant nucleic acid molecule” belong to one kind of "polynucleotide”.
- the term "recombinant nucleic acid molecule” refers to a polynucleotide having sequences that are not linked together in nature.
- the recombinant polynucleotide can be included in a suitable vector, and the vector can be used for transformation into a suitable host cell.
- the polynucleotides are then expressed in recombinant host cells to produce, for example, "recombinant polypeptides,” “recombinant proteins,” “fusion proteins,” and the like.
- linker and “linker” are used interchangeably, which are capable of linking the same or different polypeptides or amino acids.
- Linking peptides include flexible linking peptides and rigid linking peptides.
- the connecting peptide is a flexible connecting peptide.
- high stringency conditions refers to 5X SSPE (saline sodium phosphate EDTA) at 42°C for probes of at least 100 nucleotides in length following standard Southern blotting procedures , 0.3% SDS, 200 ⁇ g/ml sheared and denatured salmon sperm DNA, and 50% formamide prehybridized and hybridized for 12 to 24 hours. Finally the carrier material was washed three times for 15 min each at 65°C using 2X SSC, 0.2% SDS.
- 5X SSPE saline sodium phosphate EDTA
- very high stringency conditions refers to 5X SSPE (saline sodium phosphate EDTA) at 42°C for probes of at least 100 nucleotides in length following standard Southern blotting procedures ), 0.3% SDS, 200 ⁇ g/ml sheared and denatured salmon sperm DNA, and 50% formamide and prehybridized for 12 to 24 hours. Finally the carrier material was washed three times for 15 min each at 70°C using 2X SSC, 0.2% SDS.
- 5X SSPE saline sodium phosphate EDTA
- chimeric peptide-modified SIS membrane and “chimeric peptide-modified GBR membrane” have the same meaning and can be used interchangeably.
- SEQ ID NO: 1 shows the amino acid sequence of the P1 polypeptide (TKKTLRT+linker-1+Hst8)
- SEQ ID NO: 2 shows the amino acid sequence of the P2 polypeptide (TKKTLRT+linker-1+JH8195);
- SEQ ID NO: 3 shows the amino acid sequence of the P3 polypeptide (KELNLVY+linker-1+Hst8);
- SEQ ID NO: 4 shows the amino acid sequence of the P4 polypeptide (KELNLVY+linker-1+JH8195);
- SEQ ID NO: 5 shows the amino acid sequence of the P5 polypeptide (TKKTLRT+linker-1+Hst7)
- SEQ ID NO: 6 shows the amino acid sequence of the P6 polypeptide (TKKTLRT+linker-1+JH8944);
- SEQ ID NO: 7 shows the amino acid sequence of the P7 polypeptide (KELNLVY+linker-1+Hst7)
- SEQ ID NO: 8 shows the amino acid sequence of the P8 polypeptide (KELNLVY+linker-1+JH8944);
- SEQ ID NO: 9 shows the amino acid sequence of the P9 polypeptide (TKKTLRT+linker-1+Hst1);
- SEQ ID NO: 10 shows the amino acid sequence of the P10 polypeptide (TKKTLRT+linker-1+JH8194);
- SEQ ID NO: 11 shows the amino acid sequence of the P11 polypeptide (KELNLVY+linker-1+Hst1);
- SEQ ID NO: 12 shows the amino acid sequence of the P12 polypeptide (KELNLVY+linker-1+JH8194);
- SEQ ID NO: 13 shows the amino acid sequence of the P13 polypeptide (TKKTLRT+linker-2+Hst1);
- SEQ ID NO: 14 shows the amino acid sequence of the P14 polypeptide (TKKTLRT+linker-2+JH8194);
- SEQ ID NO: 15 shows the amino acid sequence of the P15 polypeptide (KELNLVY+linker-2+Hst1);
- SEQ ID NO: 16 shows the amino acid sequence of the P16 polypeptide (KELNLVY+linker-2+JH8194).
- the preparation method of SIS membrane is as follows:
- the small intestinal submucosa (Small Intestinal Submucosa, SIS) material was taken for primary treatment.
- the aforementioned primary treatment in the present disclosure includes the steps of dividing, washing and/or virus inactivating the SIS material.
- the virus can be inactivated by a low-concentration peracetic acid-ethanol solution method.
- This step can be carried out in an ultrasonic cleaner, wherein the volume percentage of peracetic acid can be 0.05 to 0.2%, and the ultrasonic oscillation frequency can be 30 ⁇ 600rpm, ultrasonic frequency can be 20 ⁇ 80KHZ, temperature range is 4 ⁇ 40°C, then wash in water or phosphate buffer.
- the immunogen can be removed from the small intestinal submucosa material treated in step (i) using methods including physical, chemical and/or biological methods. Including freeze-thaw method, high and low permeability method, acid-base dissolution method, detergent method, enzyme method and other methods for removal. Immunogen removal can also be performed in a combination of methods. Ultrasound-assisted means are preferred, for example in an ultrasonic cleaner.
- a decellularized solution such as an alkaline solution, a salt solution or an enzyme-containing solution, is injected into the washing tank for treatment. The processed material is cleaned.
- the small intestinal submucosa material treated in step (ii) can be used as the membrane material of the chimeric peptide-modified SIS membrane.
- the thickness and strength of the membrane material can also be increased by laminating the aforementioned (ii)-treated small intestinal submucosa material.
- the aforementioned laminated materials can also be dried.
- the drying process includes freeze-drying and/or normal temperature drying process.
- the preparation method of the chimeric peptide-modified SIS membrane is as follows:
- SIS membranes were cut into circles matching the different plate sizes and immersed in low (50 ⁇ M), medium (100 ⁇ M) and high (200 ⁇ M) chimeric peptide solutions for 10 min. Subsequently, the membranes were removed and frozen overnight, then lyophilized to generate chimeric peptide modified SIS membranes (pSIS).
- pSIS chimeric peptide modified SIS membranes
- L-pSIS is used to represent the SIS membrane soaked with the low concentration chimeric peptide solution
- M-pSIS is used to represent the SIS membrane soaked with the medium concentration chimeric peptide solution
- H-pSIS is used to represent the high concentration SIS membrane.
- SIS membrane soaked in chimeric peptide solution The chimeric peptide solutions of 50 ⁇ M, 100 ⁇ M and 200 ⁇ M were used as illustrative controls in this process, and the concentration of the chimeric peptide solution was not limited.
- the chimeric peptide can also be modified to the surface of the SIS membrane by other means, such as applying a solution containing the chimeric peptide to the surface of the SIS membrane by painting, spraying or transfer, and then drying deal with.
- PVDF polyvinylidene fluoride
- BMSCs (Bone Mesenchymal Stem Cells) or OECs were seeded on 24-well plates, cultured with 1 mL of SIS membrane, M-pSIS membrane or H-pSIS membrane extract, and then BMSCs were cultured with osteogenic induction medium . After the corresponding incubation period, BMSCs or OECs were fixed in 4% formaldehyde for 30 min, permeabilized with 0.5% Triton X-100 for 5 min and blocked in 5 mg/mL BSA solution for 1 h. Cells were then incubated with primary antibodies (Abeam, UK) at a 1:200 dilution for 2 h at 37°C.
- primary antibodies Abeam, UK
- the experimental techniques and experimental methods used in the present embodiment are conventional technical methods, such as the experimental methods that do not specify specific conditions in the following examples, usually according to conventional conditions such as people such as Sambrook, molecular cloning: experiment The conditions described in the Laboratory Manual (New York: Cold Spring Harbor Laboratory Press, 1989), or as suggested by the manufacturer. Materials, reagents, etc. used in the examples can be obtained through regular commercial channels unless otherwise specified.
- the present disclosure designed a combination of chimeric peptides comprising sets of different functions.
- the chimeric polypeptides involved in the present disclosure were synthesized by Jill biochemical Shanghai Co., Ltd, China.
- Control-1 collagen binding peptide (type I: TKKTLRT; type III: KELNLVY); flexible Linker: GGGGSGGGGS; functional peptide: Hst8/JH8195.
- the sequences of the four chimeric peptides in Control-1 are shown in Table 1.
- Control-2 collagen binding peptide (type I: TKKTLRT; type III: KELNLVY); flexible Linker: GGGGSGGGGS; functional peptide: Hst7/JH8944.
- the sequences of the four chimeric peptides in Control-2 are shown in Table 2.
- pSIS-1 collagen-binding peptide (type I: TKKTLRT; type III: KELNLVY); flexible Linker: GGGGSGGGGS; functional peptide: Hst1/JH8194; the sequences of the four chimeric peptides in pSIS-1 are shown in Table 3. Among them, Hst1 has a certain degree of healing activity, and JH8194 has a certain degree of osteogenic activity and antibacterial activity.
- pSIS-2 collagen binding peptide (type I: TKKTLRT; type III: KELNLVY); rigid Linker: EAAAAKEAAAK; functional peptide: Hst1/JH8194; the sequences of the four chimeric peptides in pSIS-2 are shown in Table 4.
- pSIS in Example 2 corresponds to pSIS-1 in Example 1.
- Chimeric peptides were synthesized to 95% purity by Fmoc solid-phase peptide synthesis (Jill biochemical Shanghai Co., Ltd, China) according to the sequences in Table 3.
- P9, P10 were labeled with FITC and P11, P12 were labeled with RB for CLSM.
- the secondary structure was analyzed with the help of the software PSIPRED.
- the tertiary structure was predicted with the aid of the protein analysis software Robetta and visualized with the aid of VMD.
- the structures of P9, P10, P11 and P12 were predicted by PSIPRED and Robetta.
- the chimeric peptide consists of three parts: P9 and P10: TKKTLRT (type I collagen bound to the SIS membrane), Hst1/JH8194 (functioning as antibacterial, osteogenic and healing promoting abilities) and GGGGSGGGGS (connecting the first two parts), P11 and P12: KELNLVY (type III collagen bound to SIS membrane), Hst1/JH8194 and GGGGSGGGGS.
- FIG. 1A shows the design details and structures of the collagen chimeric peptides P9 and P10.
- Figure IB shows the design details and structures of the collagen chimeric peptides P11 and P12.
- the four chimeric peptides consisted of collagen-binding peptides (TKKTLRT or KELNLVY), functional peptides (Hst1 or JH8194) and flexible linkers (GGGGSGGGGS).
- GGGGS is the most commonly used flexible linker composed of glycine (Gly) and serine (Ser) residues, which ensures flexibility and mobility for connecting functional domains.
- the copy number of "GGGGS” is preferably 2 to achieve proper separation of functional domains and avoid mutual interference.
- pSIS in Example 3 corresponds to pSIS-1 in Example 1.
- SIS membranes were cut into circles matching the different plate sizes and immersed in low (50 ⁇ M), medium (100 ⁇ M) and high (200 ⁇ M) chimeric peptide solutions for 10 min. Subsequently, the membranes were removed and frozen overnight, then lyophilized to generate chimeric peptide modified SIS membranes (pSIS). The samples were labeled as L-pSIS group, M-pSIS group and H-pSIS group. After sputtering with gold, the surface morphology of the SIS and pSIS films was observed by SEM (Gemini 300, Zeiss, Germany).
- BMSCs or OECs in 1000 ⁇ L of Dulbecco’s modified Eagle’s medium (DMEM, Gibco, USA) containing 10% fetal bovine serum (FBS, Gibco, USA) were seeded onto 24-well plates and placed on Incubate at 37°C, 5% CO 2 .
- DMEM Dulbecco’s modified Eagle’s medium
- FBS fetal bovine serum
- 100 ⁇ L of CCK-8 solution (Solarbio, China) was added to each sample. Relative cell viability was determined by measuring the absorbance (OD) at 450 nm after 4 h incubation at 37°C, 5% CO2 .
- Figure 2A shows a schematic diagram of a pSIS membrane.
- Figure 2B shows the binding of chimeric peptides observed by CLSM. Among them, P9 and P10 were labeled with rhodamine B (RB), and P11 and P12 were labeled with fluorescein isothiocyanate (FITC).
- Figure 2C shows the surface morphology of lyophilized SIS and pSIS observed by SEM.
- Figure 2D shows the proliferative capacity of BMSCs and OECs by CCK-8.
- SIS membranes were immersed in 50 ⁇ M (L-pSIS), 100 ⁇ M (M-pSIS) and 200 ⁇ M (H-pSIS) chimeric peptide solutions to observe the binding of chimeric peptides on the surface of SIS membranes, we used co- Focused Laser Scanning Microscopy (CLSM) and Scanning Electron Microscopy (SEM) were used for detection. Fluorescent labeling on the SIS membrane increased with increasing concentration (Fig. 2B), indicating more chimeric peptide binding. In particular, more than 85% of the fluorescent area of the SIS membrane (H-pSIS) achieved efficient binding after soaking with 200 ⁇ M chimeric peptide solution.
- CLSM co- Focused Laser Scanning Microscopy
- SEM Scanning Electron Microscopy
- Biocompatibility the interaction between material and host, is an essential characteristic of materials to ensure patient safety during application.
- the effect of pSIS on cell proliferation over time was investigated by CCK-8. It showed that BMSCs and OECs proliferated logarithmically on the surface of pSIS (Fig. 2D).
- the proliferation ability of BMSCs on the H-pSIS membrane was significantly lower than that of other groups, but as time went on, the proliferation ability increased and was not statistically different from other groups. It was found that pSIS had no significant effect on the proliferation of BMSCs or OECs, providing assurance for their further application. Therefore, considering the results of CLSM, SEM and CCK-8, we selected M-pSIS and H-pSIS with higher binding rate of chimeric peptides for further study in subsequent experiments.
- pSIS in Example 4 corresponds to pSIS-1 in Example 1.
- Streptococcus sanguis (ATCC 10556) and Streptococcus griffin (ATCC 51656) were grown on brain heart infusion (BHI) agar plates for 18 h at 37°C. The bacteria were then resuspended at 1 ⁇ 10 7 CFU/ml (CFU, colony forming unit) as the main inoculum. In a 100 mm bacterial petri dish, inoculate 20 ml of BHI solution mixed with 100 ⁇ l of bacterial solution. After solidification, the SIS, M-pSIS, and H-pSIS membranes were centered and blank wells were used as controls. After 24 hours, the inhibition zone was observed and photographed.
- BHI brain heart infusion
- Figure 3A shows the inhibition zone of S. sanguis and S. gordonii (upper left: blank control; upper right: SIS; lower left: M-pSIS; lower right: H-pSIS) .
- Figure 3B shows CLSM images stained after culturing bacteria in blank wells, SIS, M-pSIS or H-pSIS for 24 hours.
- JH8194 is a derived peptide of oral antimicrobial peptide Hst5.
- AMP and its derivatives can exchange divalent cations (eg, Mg 2+ and Ca 2+ ) on bacterial membranes by an "ion exchange mechanism" to destabilize the cytoplasmic membrane, leading to cell death.
- positively charged AMPs can adsorb on negatively charged cytoplasmic membranes through electrostatic interactions and intercalate into phospholipid bilayers to form pores or even larger defects in the membranes, ultimately leading to cytoplasmic leakage and bacterial death.
- Example 5 The effect of pSIS on the expression of osteogenesis-related factors in BMSCs
- pSIS in Example 5 corresponds to pSIS-1 in Example 1.
- SIS membrane, M-pSIS membrane or H-pSIS membrane and 500 ⁇ L of medium (2% FBS) were placed in the lower compartment of the transwell plate, and blank wells with medium only were used as controls. Then, 1 ⁇ 10 4 OECs in 500 ⁇ L medium (2% FBS) were seeded onto transwell inserts (Thermo Fisher Scientific, USA). After 24 h, the medium in the transwell insert was carefully removed. Cells were fixed with 4% paraformaldehyde for 30 min, permeabilized with 0.01% Triton X-100 (Sigma-Aldrich, USA) for 5 min and stained with 1% crystal violet (Sigma-Aldrich, USA). Unmigrated cells were removed by wiping gently with a cotton swab. Then, the migrated cells were observed and imaged with an inverted fluorescence microscope (Olympus IX71, Japan).
- the in vitro osteogenic activity of pSIS was determined as follows:
- the rats were euthanized. Bone defects were removed and fixed in 4% paraformaldehyde for 24 h. The samples were then scanned with Micro-CT (SkyScan 1276, Germany) for standardized reconstruction and evaluation of new bone formation. Next, the samples were decalcified with rapid decalcification solution (Rapid Cal Immuno, ZS-Bio, China) for 7d. After dehydration and embedding in paraffin, the samples were sectioned at a thickness of 5 ⁇ m. H&E and Masson's trichrome staining (Solarbio, China) was used for histological analysis.
- Osteogenic induction medium was prepared containing 10 mM ⁇ -glycerophosphate (Sigma, USA), 100 nM dexamethasone (Sigma, USA) and 50 ⁇ M ascorbate (Sigma, USA). 1 ⁇ 10 5 BMSCs were seeded on sterile SIS membranes, M-pSIS membranes and H-pSIS membranes in 6-well plates, cultured with medium for 3 days, and then replaced with osteogenic induction medium. After 7 days, 14 days and 21 days, the expression of BMP2, RUNX2, ALP and OPN was detected.
- Figure 4A shows the results of qPCR analysis of osteogenesis-related genes (* indicates p ⁇ 0.05).
- Figure 4B shows the results of immunoblot analysis of osteogenesis-related proteins.
- Figure 4C shows the quantitative analysis results of immunoblot analysis (* indicates p ⁇ 0.05).
- Figure 4D shows the results of immunofluorescence analysis of the localization of osteogenesis-related proteins in BMSCs (BMP2, RUNX2, ALP, OPN and nucleus).
- Osteogenesis is the key to GBR technology.
- the effect of pSIS on the expression of osteogenesis-related factors by BMSCs was also evaluated (FIG. 4A-FIG. 4D).
- Bone morphogenetic protein-2 (BMP2), Runt-associated transcription factor 2 (RUNX2), alkaline phosphatase (ALP) and osteopontin (OPN) are classical osteogenic factors.
- BMP2 Bone morphogenetic protein-2
- RUNX2 Runt-associated transcription factor 2
- ALP alkaline phosphatase
- OPN osteopontin
- the mRNA levels of the above osteogenic factors were significantly higher in the pSIS group than in the blank control and SIS.
- immunoblot analysis also confirmed this trend (Fig. 4B and Fig. 4C), indicating that chimeric peptide-modified SIS membranes could further promote the expression of osteogenesis-related factors.
- Example 6 Effects of pSIS on OEC migration and possible signaling pathways
- pSIS in Example 6 corresponds to pSIS-1 in Example 1.
- SD rats were anesthetized by inhalation of isoflurane and fixed in the prone position. After removing the back hair, four symmetrical circular marks were formed on both sides of the spine by using a drill (2 cm in diameter). Along the circular marks, the entire skin was cut with a scalpel and divided into the following groups: Blank, SIS membrane group, Bio-Gide membrane group, and H-pSIS membrane group, and the membranes were sutured and Subcutaneous tissue fixation. Each rat was housed separately to avoid scratching each other. The specific methods of H&E, Masson's trichrome staining and immunohistochemistry were the same as previously described. The expression of ITG- ⁇ 1 and ITG- ⁇ 3 was detected by immunohistochemistry.
- Figure 5A shows the results of the Transwell assay to detect cell migration ability.
- Figure 5B shows the results of the analysis of migrated cells (* indicates p ⁇ 0.05).
- Figure 5C shows the results of differentially expressed genes screened by RNA-seq.
- Figure 5D shows a Venn diagram of different genes.
- Figure 5E shows the results of qPCR analysis of migration-related genes.
- Figure 5F shows the results of Western blot analysis of migration-related proteins.
- Figure 5G shows the results of quantitative analysis of Western blot analysis (* indicates p ⁇ 0.05).
- Figure 5H shows the results of immunofluorescence analysis of migration-related protein localization in OECs (ITG-[alpha]3, ITG-[beta]1 and nucleus).
- the wound healing process in GBR involves cell proliferation and migration, wound contraction and angiogenesis, collagen deposition and remodeling.
- Cell migration which consists of a multi-step process, is required for healing repair.
- the sequence "SHREFPFYGDYGS" of Hst1 contains the minimum elements necessary to promote cell migration, which can promote the migration of oral keratinocytes, oral epithelial cells and gingival fibroblasts.
- the effect of pSIS on the migration of OECs was examined by Transwell experiments (Fig. 5A and Fig. 5B). SIS and pSIS can promote the migration of OECs, among which H-pSIS has the strongest ability, indicating that the chimeric peptide-modified SIS membrane can further promote cell migration, and may contribute to the healing of soft tissue.
- RNA-seq a novel high-throughput sequencing method, RNA-seq, was used to analyze the differentially expressed genes of OECs cultured on SIS, M-pSIS, and H-pSIS for 24 h (Fig. 5C).
- 790 genes were up-regulated and 833 genes were down-regulated in the M-pSIS group
- 756 genes were up-regulated and 735 genes were down-regulated in the H-pSIS group.
- the Venn diagram shows 844 genes with the same trend.
- Integrin ⁇ 3 ⁇ 1 is closely related to cell migration and wound healing. It can promote re-epithelialization by accelerating keratinocyte migration to assist in epithelial wound healing. In the GBR region, integrin ⁇ 3 ⁇ 1 can bind to unprocessed laminin 5 and then mediate the migration of connected epithelial cells. Therefore, the expression of ITG- ⁇ 3 and ITG- ⁇ 1 in OECs was detected.
- Example 7 Evaluation of the osteogenic capacity of pSIS in vivo
- pSIS in Example 7 corresponds to pSIS-1 in Example 1.
- Figure 6A shows three-dimensional reconstructions and sagittal images of bone defects covered with SIS, Bio-Gide and H-pSIS membranes.
- Figure 6B shows BV/TV results for different groups of bone defects (* indicates p ⁇ 0.05).
- Figure 6C shows H&E staining results.
- Figure 6D shows the results of Masson's trichrome staining.
- Figure 6E shows the results of immunohistochemical analysis of OCN and COL1 expression.
- FIG. 6A-Fig. 6E A critical size defect (CSD) model of a non-self-healing 8 mm circular defect was made in rats to evaluate bone regeneration.
- Bio-Gide membrane commercially purchased
- CT microcomputed tomography
- H&E hematoxylin-eosin
- Masson's trichrome staining to evaluate collagen and new bone tissue growth and lymphocyte infiltration.
- Collagen is an important building block of bones.
- the ECM secreted by osteoblasts, including collagen type I, can transition from initially amorphous and non-crystalline to more crystalline, gradually leading to osteogenesis.
- mineralization is the main process by which osteoblasts promote bone formation.
- Collagen acts as a template and can also initiate and propagate mineralization. Therefore, collagen content is closely related to bone formation.
- the defect area of the blank control group was mainly composed of fibrous tissue, with less collagen, and no obvious signs of new bone formation.
- the bone defect areas in the SIS, Bio-Gide and H-pSIS groups were rich in collagen, and new bone formed around the stump with little or no infiltration of fibrous connective tissue. Meanwhile, lymphocyte infiltration was low in all groups, indicating no inflammatory response (Fig. 6C).
- the expression of osteocalcin (OCN) and collagen type I (COL1) was detected by immunohistochemistry (Fig. 6E). Twelve weeks after the operation, no obvious OCN and COLI staining were found in the control group, but they were highly expressed in the SIS, Bio-Gide and H-pSIS groups, and the H-pSIS group was the highest.
- H-pSIS could effectively prevent fibrous connective tissue from growing into the defect area and provide space for bone formation.
- the excellent bone regeneration ability of pSIS helps to solve the problem of alveolar bone loss and insufficient bone in the dental implant area caused by periodontitis or tooth loss.
- its unique anti-infective ability helps to resist and prevent tissue inflammation and infection, thereby repairing infectious bone defects, which is expected to solve the clinical problem of repairing infection-related bone defects.
- Example 8 Evaluation of the ability of pSIS to promote healing in vivo
- pSIS in Example 8 corresponds to pSIS-1 in Example 1.
- Figure 7A shows H&E staining results (line segment: length of non-epithelial wound).
- Figure 7B shows the length of neo-epithelialization (* indicates p ⁇ 0.05).
- Figure 7C shows the results of Masson's trichrome staining (line segment: length of non-collagenous fibers of the wound).
- Figure 7D shows collagen fiber length (* indicates p ⁇ 0.05).
- Figure 7E shows the results of immunohistochemical analysis of ITG- ⁇ 3 and ITG- ⁇ 1 expression.
- Fig. 7A-Fig. 7D histological analysis was performed by H&E and Masson's trichrome staining.
- Re-epithelialization an essential feature of wound healing, is associated with directed migration of keratinocytes.
- pSIS membranes can promote re-epithelialization by promoting cell migration, which contributes to rapid wound healing.
- Example 9 Comparison of osteogenic, inflammatory and healing effects of SIS films modified with different chimeric peptides
- Control-1 is a mixture of chimeric peptides represented by P1-P4;
- Control-2 is a mixture of chimeric peptides represented by P5-P8;
- pSIS-1 is a mixture of chimeric peptides represented by P9-P12 A mixture of chimeric peptides;
- pSIS-2 is a mixture of chimeric peptides represented by P13-P16.
- pSIS-1 has better osteogenic effect, anti-inflammatory effect and healing effect.
- chimeric peptide-modified SIS film developed in the present disclosure can simultaneously exert antibacterial effects, promote soft tissue healing and bone regeneration, and greatly enrich the performance of the GBR film.
- the present disclosure proves that the chimeric peptide-modified SIS film (ie, the GBR film) can For the clinical treatment of infected bone defects.
Landscapes
- Health & Medical Sciences (AREA)
- Chemical & Material Sciences (AREA)
- Life Sciences & Earth Sciences (AREA)
- Medicinal Chemistry (AREA)
- General Health & Medical Sciences (AREA)
- Epidemiology (AREA)
- Public Health (AREA)
- Dermatology (AREA)
- Oral & Maxillofacial Surgery (AREA)
- Transplantation (AREA)
- Veterinary Medicine (AREA)
- Animal Behavior & Ethology (AREA)
- Biomedical Technology (AREA)
- Engineering & Computer Science (AREA)
- Molecular Biology (AREA)
- Biochemistry (AREA)
- Organic Chemistry (AREA)
- Botany (AREA)
- Chemical Kinetics & Catalysis (AREA)
- Biophysics (AREA)
- Genetics & Genomics (AREA)
- Proteomics, Peptides & Aminoacids (AREA)
- Peptides Or Proteins (AREA)
Abstract
一种嵌合肽修饰的SIS膜、其制备方法及应用。利用嵌合肽修饰的SIS膜促进了ITG-α3、ITG-β1、BMP2、RUNX2、ALP和OPN的表达,并抑制了格氏链球菌和血链球菌的生长,从而使SIS膜发挥抗菌、成骨和促进愈合的生物功能。所述SIS膜(即嵌合肽修饰的GBR膜)能够用于感染性骨缺损的临床治疗。
Description
本公开属于生物医用材料领域,具体涉及一种结合嵌合肽的SIS膜、其制备方法及应用。
由创伤、肿瘤、炎症和其它原因引起的骨缺损仍然是目前骨重建中不可弥补的障碍。牙周炎和种植体周围炎是由于菌斑生物膜导致的牙槽骨质丢失的常见原因,最终可能导致牙齿或种植体丢失以及牙齿种植体区域的骨骼不足。认为引导骨再生(Guided Bone Regeneration,GBR)技术是重建骨缺损最常用的方法之一。它需要屏障膜来防止上皮细胞和纤维结缔组织生长到缺损区域中,从而确保成骨细胞可以填充骨缺损空间以获得期望的骨再生。GBR膜的性能在GBR技术的应用中起着重要作用,它与GBR手术后的细菌感染的预防、伤口愈合和骨再生密切相关。
细胞外基质(Extracellular Matrix,ECM)材料是由胶原(主要是I型和III型)和各种糖蛋白组成的非细胞生物网络,近年来已成为组织工程支架的热点。小肠粘膜下层(Small Intestinal Submucosa,SIS)由于其来源广泛且易于获得因而是最常用的ECM材料。SIS具有许多优异的性能,包括部位特异性组织再生能力、低免疫原性、优异的机械性能、以及包含多种细胞因子。它已在许多类型的组织中用作支架材料,并且获得了令人满意的结果。然而,由牙周炎、种植体周围炎和根尖周炎引起的感染性骨缺损是GBR的难题。尽管SIS膜具有优异的机械性能和生物相容性,但是其抗菌、成骨和促进愈合的能力受到限制。为了解决这些限制,希望找到合适的方式来用于SIS的修饰。
作为天然免疫的重要部分的抗菌肽(Antimicrobial Peptides,AMP)具有广谱、强大且稳定的抗菌活性以及良好的生物相容性,已成为新一代潜在的抗菌药物。组胺素(Histatins(HST))是一类由人类主要唾液腺分泌的AMP。由于阳离子性和弱的两亲性,HST在温暖和潮湿的口腔环境中表现出广谱抗菌活性。Hst5是HST的主要成员,它可以通过IKK/NFκB通路抵抗牙龈卟啉单胞菌(Porphyromonas gingivalis(Pg))的LPS(Lipopolysaccharide),并且抑制由Pg外膜蛋白诱导的IL-6和IL-8的表达。以前的研究表明,Hst5的衍生肽JH8194也具有良好的抗菌活性,可以有效地预防种植体周围炎和种植体周围粘膜炎。除了抗菌活性,JH8194还具有一定的促进骨形成的能力。已经发现JH8194可以在抑制Pg的生物膜形成的同时刺激MC3T3-E1中Runx2、Opn和Alp的表达,从而使细胞可以分化为成骨细胞。此外,JH8194可以增加种植体周围的成熟小梁骨的形成并且增强骨整合,这有利于种植体的长期稳定性。因此,JH8194是用于SIS膜的抗菌和成骨修饰的有效药物。
令人惊讶的是,HST的另一个成员Hst1在促进愈合方面表现出优异的能力。Hst1可以促进成纤维细胞、上皮细胞和内皮细胞的迁移或粘附,以及引导伤口区域的表皮细胞再生和血管再生。一些研究发现,Hst1与G蛋白偶联受体(G Protein-Coupled Receptors,GPCR)结合并且激活G蛋白,随后激活下游未知激酶并且诱导细胞外信号调节激酶(Extracellular Regulated Protein Kinases,Erk)1/2/丝裂原活化蛋白(Microtubule Associated Protein,MAP)磷酸化,最终促进细胞增殖和迁移。因此,在JH8194的基础上负载Hst1可以增强SIS的促进愈合的功能,但仍需探索具体的机制。尽管有这些优点,但是,仍然需要提供一种具有良好抗菌作用,促进软组织愈合和/或骨再生作用的GBR膜。
发明内容
发明要解决的问题
基于现有技术存在的缺陷,本公开提供了一种嵌合肽修饰的SIS膜可同时发挥抗菌作用,促进软组织愈合和骨再生的功能,极大地丰富了GBR膜的性能,本公开证明了嵌合肽修饰的SIS膜(即GBR膜)可以用于感染性骨缺损的临床治疗。
用于解决问题的方案
本公开记载了如下技术方案。
(1)一种嵌合肽修饰的SIS膜,其中,所述嵌合肽的序列包含由如下序列所示的序列组成的组中 的至少一种:
(i)如SEQ ID NO:9所示的序列、如SEQ ID NO:10所示的序列、如SEQ ID NO:11所示的序列、如SEQ ID NO:12所示的序列组成的组;
(ii)和(i)所示的序列相比,存在保守置换的序列。
在一个实施方式中,所述(1)中的嵌合肽的序列包含和如SEQ ID NO:9所示的序列、如SEQ ID NO:10所示的序列、如SEQ ID NO:11所示的序列、如SEQ ID NO:12所示的序列相比,具有至少80%以上同一性的序列。
在一个具体的实施方式中,所述(1)中的嵌合肽的序列包含和如SEQ ID NO:9所示的序列、如SEQ ID NO:10所示的序列、如SEQ ID NO:11所示的序列、如SEQ ID NO:12所示的序列相比,具有80%、81%、82%、83%、84%、85%、86%、87%、88%、89%、90%、91%、92%、93%、94%、95%、96%、97%、98%或99%以上的同一性的序列。
在另一个具体的实施方式中,所述(1)中的嵌合肽的序列包含和如SEQ ID NO:9所示的序列、如SEQ ID NO:10所示的序列、如SEQ ID NO:11所示的序列、如SEQ ID NO:12所示的序列相比,具有80%、81%、82%、83%、84%、85%、86%、87%、88%、89%、90%、91%、92%、93%、94%、95%、96%、97%、98%或99%以上的同一性的序列;并且,其和如SEQ ID NO:9所示的序列、如SEQ ID NO:10所示的序列、如SEQ ID NO:11所示的序列、如SEQ ID NO:12所示的序列相比,仅存在保守置换。
(2)根据(1)所述的嵌合肽修饰的SIS膜,其中,所述嵌合肽的序列由如下序列组成的组中的至少一种组成:
(i)如SEQ ID NO:9所示的序列、如SEQ ID NO:10所示的序列、如SEQ ID NO:11所示的序列、如SEQ ID NO:12所示的序列组成的组;
(ii)和(i)所示的序列相比,存在保守置换的序列。
在一个实施方式中,所述(2)中的嵌合肽的序列和如SEQ ID NO:9所示的序列、如SEQ ID NO:10所示的序列、如SEQ ID NO:11所示的序列、如SEQ ID NO:12所示的序列相比,具有至少80%以上同一性。
在一个具体的实施方式中,所述(2)中的嵌合肽的序列和如SEQ ID NO:9所示的序列、如SEQ ID NO:10所示的序列、如SEQ ID NO:11所示的序列、如SEQ ID NO:12所示的序列相比,具有80%、81%、82%、83%、84%、85%、86%、87%、88%、89%、90%、91%、92%、93%、94%、95%、96%、97%、98%或99%以上的同一性。
在另一个具体的实施方式中,所述(2)中的嵌合肽的序列和如SEQ ID NO:9所示的序列、如SEQ ID NO:10所示的序列、如SEQ ID NO:11所示的序列、如SEQ ID NO:12所示的序列相比,具有80%、81%、82%、83%、84%、85%、86%、87%、88%、89%、90%、91%、92%、93%、94%、95%、96%、97%、98%或99%以上的同一性;并且,其和如SEQ ID NO:9所示的序列、如SEQ ID NO:10所示的序列、如SEQ ID NO:11所示的序列、如SEQ ID NO:12所示的序列相比,仅存在保守置换。
(3)根据(1)-(2)任一项所述的嵌合肽修饰的SIS膜,其中,所述修饰的方法包括以下步骤:
(a)将所述嵌合肽溶解至溶剂中,得到含有嵌合肽的溶解液;
(b)将步骤(a)得到的溶解液施加至所述SIS膜的表面;优选的,将所述SIS膜浸入步骤(a)得到的溶解液中;
(c)干燥表面带有所述溶解液的所述SIS膜。
(4)根据(1)-(3)任一项所述的嵌合肽修饰的SIS膜,其中,所述SIS膜的制备方法包括以下步骤:
(i)取小肠粘膜下层材料进行初处理;
(ii)将经步骤(i)得到的小肠粘膜下层材料进行免疫原去除处理。
(5)根据(4)所述的嵌合肽修饰的SIS膜,其中,所述SIS膜的制备方法还包括以下步骤:
(iii)将经步骤(ii)得到的小肠粘膜下层材料进行层叠;
(iv)将层叠后的小肠粘膜下层材料进行干燥处理。
(6)一种嵌合肽修饰的SIS膜的制备方法,其中,所述方法包括以下步骤:
(a)将所述嵌合肽溶解至溶剂中,得到含有嵌合肽的溶解液;
(b)将步骤(a)得到的溶解液施加至所述SIS膜表面;优选的,将所述SIS膜浸入步骤(a)得到的溶解液中;
(c)干燥表面带有所述溶解液的所述SIS膜,得到所述嵌合肽修饰的SIS膜;
其中,所述嵌合肽的序列包含由如下序列所示的序列组成的组中的至少一种,或者所述嵌合肽的序列由如下序列组成的组中的至少一种组成:
(i)如SEQ ID NO:9所示的序列、如SEQ ID NO:10所示的序列、如SEQ ID NO:11所示的序列、如SEQ ID NO:12所示的序列组成的组;
(ii)和(i)所示的序列相比,存在保守置换的序列。
在一个实施方式中,所述(6)中的嵌合肽的序列包含和如SEQ ID NO:9所示的序列、如SEQ ID NO:10所示的序列、如SEQ ID NO:11所示的序列、如SEQ ID NO:12所示的序列相比,具有至少80%以上同一性的序列;或者,所述(6)中的嵌合肽的序列和如SEQ ID NO:9所示的序列、如SEQ ID NO:10所示的序列、如SEQ ID NO:11所示的序列、如SEQ ID NO:12所示的序列相比,具有至少80%以上同一性。
在一个具体的实施方式中,所述(6)中的嵌合肽的序列包含和如SEQ ID NO:9所示的序列、如SEQ ID NO:10所示的序列、如SEQ ID NO:11所示的序列、如SEQ ID NO:12所示的序列相比,具有80%、81%、82%、83%、84%、85%、86%、87%、88%、89%、90%、91%、92%、93%、94%、95%、96%、97%、98%或99%以上的同一性的序列;或者,所述(6)中的嵌合肽的序列和如SEQ ID NO:9所示的序列、如SEQ ID NO:10所示的序列、如SEQ ID NO:11所示的序列、如SEQ ID NO:12所示的序列相比,具有80%、81%、82%、83%、84%、85%、86%、87%、88%、89%、90%、91%、92%、93%、94%、95%、96%、97%、98%或99%以上的同一性。
在另一个具体的实施方式中,所述(6)中的嵌合肽的序列包含和如SEQ ID NO:9所示的序列、如SEQ ID NO:10所示的序列、如SEQ ID NO:11所示的序列、如SEQ ID NO:12所示的序列相比,具有80%、81%、82%、83%、84%、85%、86%、87%、88%、89%、90%、91%、92%、93%、94%、95%、96%、97%、98%或99%以上的同一性的序列;并且,其和如SEQ ID NO:9所示的序列、如SEQ ID NO:10所示的序列、如SEQ ID NO:11所示的序列、如SEQ ID NO:12所示的序列相比,仅存在保守置换。
在另一个具体的实施方式中,所述(6)中的嵌合肽的序列和如SEQ ID NO:9所示的序列、如SEQ ID NO:10所示的序列、如SEQ ID NO:11所示的序列、如SEQ ID NO:12所示的序列相比,具有80%、81%、82%、83%、84%、85%、86%、87%、88%、89%、90%、91%、92%、93%、94%、95%、96%、97%、98%或99%以上的同一性;并且,其和如SEQ ID NO:9所示的序列、如SEQ ID NO:10所示的序列、如SEQ ID NO:11所示的序列、如SEQ ID NO:12所示的序列相比,仅存在保守置换。
(7)根据(6)所述的嵌合肽修饰的SIS膜的制备方法,其中,所述SIS膜的制备方法包括以下步骤:
(i)取小肠粘膜下层材料进行初处理;
(ii)将经步骤(i)得到的小肠粘膜下层材料进行免疫原去除处理。
(8)根据(7)所述的嵌合肽修饰的SIS膜的制备方法,其中,所述SIS膜的制备方法还包括以下步骤:
(iii)将经步骤(ii)得到的小肠粘膜下层材料进行层叠;
(iv)将层叠后的小肠粘膜下层材料进行干燥处理。
(9)根据(1)-(5)任一项所述的嵌合肽修饰的SIS膜或根据(6)-(8)任一项所述的嵌合肽修饰的SIS膜的制备方法得到的嵌合肽修饰的SIS膜在如下(a)-(d)至少一种中的用途:
(a)作为或制备抗菌的生物材料;
(b)作为或制备成骨的生物材料;
(c)作为或制备促进愈合的生物材料;
(d)作为或制备治疗感染性骨缺损的生物材料。
(10)一种治疗感染性骨缺损的方法,其中,所述方法包括向受试者施用如(1)-(5)任一项所述的嵌合肽修饰的SIS膜或根据(6)-(8)任一项所述的嵌合肽修饰的SIS膜的制备方法得到的嵌合肽修饰的SIS膜的步骤。
发明的效果
在一个实施方式中,本公开成功开发了利用嵌合肽修饰的SIS膜,用于制备嵌合肽修饰的GBR膜。
在一个具体的实施方式中,本公开采用了一组抗菌、成骨和促进愈合的嵌合肽通过胶原结合序列靶向SIS膜的表面。
在一个具体的实施方式中,本公开利用嵌合肽修饰的SIS膜,促进了ITG-α3、ITG-β1、BMP2、RUNX2、ALP和OPN的表达,并抑制了格氏链球菌和血链球菌的生长,从而使SIS膜发挥抗菌、成骨和促进愈合的生物功能。
在一个具体的实施方式中,本公开采用的利用嵌合肽修饰的SIS膜(即嵌合肽修饰的GBR膜)能够用于感染性骨缺损的临床治疗。
图1A示出了胶原嵌合肽P9和P10的设计详情和结构。其中,图例为折叠链(Strand)、螺旋(Helix)和无规则卷曲(Coil)。Conf表示置信度预测(Confidence of prediction);Cart表示三态卡通图(3-state assignment cartoon);Pred表示三态预测(3-state prediction);AA表示目标序列(Target Sequence)。
图1B示出了胶原嵌合肽P11和P12的设计详情和结构。其中,图例为折叠链(Strand)、螺旋(Helix)和无规则卷曲(Coil)。Conf表示置信度预测(Confidence of prediction);Cart表示三态卡通图(3-state assignment cartoon);Pred表示三态预测(3-state prediction);AA表示目标序列(Target Sequence)。
图2A示出了pSIS膜的示意图。
图2B示出了CLSM观察的嵌合肽的结合。
图2C示出了通过SEM观察冻干SIS和pSIS的表面形态。
图2D示出了通过CCK-8的BMSC和OEC的增殖能力。
图3A示出了血链球菌和格氏链球菌的抑菌圈。
图3B示出了在空白孔、SIS、M-pSIS或H-pSIS中培养混合细菌24小时后染色的CLSM图像。
图4A示出了成骨相关基因的qPCR分析结果(*表示p<0.05)。
图4B示出了成骨相关蛋白的免疫印迹分析结果。
图4C示出了免疫印迹分析的定量分析结果(*表示p<0.05)。
图4D示出了BMSC中成骨相关蛋白定位的免疫荧光分析结果(BMP2、RUNX2、ALP、OPN和细胞核)。
图5A示出了Transwell实验检测细胞迁移能力的结果。
图5B示出了迁移细胞的分析的结果(*表示p<0.05)。
图5C示出了RNA-seq筛选的差异表达基因的结果。
图5D示出了不同基因的维恩图(Venn diagram)。
图5E示出了迁移相关基因的qPCR分析的结果。
图5F示出了迁移相关蛋白的蛋白质印迹分析的结果。
图5G示出了蛋白质印迹分析的定量分析的结果(*表示p<0.05)。
图5H示出了OEC中迁移相关蛋白定位的免疫荧光分析的结果(ITG-α3、ITG-β1和细胞核)。
图6A示出了用SIS、Bio-Gide和H-pSIS膜覆盖的骨缺损的三维重建和矢状图像。
图6B示出了不同组的骨缺损的BV/TV结果(*表示p<0.05)。
图6C示出了H&E染色结果。
图6D示出了Masson的三色染色的结果。
图6E示出了OCN和COL1表达的免疫组织化学分析结果。
图7A示出了H&E染色结果(线段:非上皮伤口的长度)。
图7B示出了新上皮化长度(*表示p<0.05)。
图7C示出了Masson的三色染色结果(线段:伤口的非胶原纤维的长度)。
图7D示出了胶原纤维长度(*表示p<0.05)。
图7E示出了ITG-α3和ITG-β1表达的免疫组织化学分析结果。
图8示出了大鼠实验在手术后1和2周后的愈合结果。
图9示出了不同嵌合肽修饰的SIS膜的成骨效果比较。
图10示出了不同嵌合肽修饰的SIS膜的抑制炎症效果比较。
图11示出了不同嵌合肽修饰的SIS膜的愈合效果比较。
定义
当在权利要求和/或说明书中与术语“包含”联用时,词语“一(a)”或“一(an)”可以指“一个”,但也可以指“一个或多个”、“至少一个”以及“一个或多于一个”。
如在权利要求和说明书中所使用的,词语“包含”、“具有”、“包括”或“含有”是指包括在内的或开放式的,并不排除额外的、未引述的元件或方法步骤。
在整个申请文件中,术语“约”表示:一个值包括测定该值所使用的装置或方法的误差的标准偏差。
虽然所公开的内容支持术语“或”的定义仅为替代物以及“和/或”,但除非明确表示仅为替代物或替代物之间相互排斥外,权利要求中的术语“或”是指“和/或”。
如本公开所使用的,术语“氨基酸突变”或“核苷酸突变”,包括“取代、重复、缺失或添加一个或多个氨基酸或核苷酸”。在本公开中,术语“突变”是指核苷酸序列或者氨基酸序列的改变。在一些实施方案中,本公开的“突变”可以选自“保守突变”、“半保守突变”、“非保守突变”。在本公开中,术语“非保守突变”或“半保守突变”可以是引起蛋白功能丧失或部分丧失的突变。术语“保守突变”是指可正常维持蛋白质的功能的突变。保守突变的代表性例子为保守置换。
如本公开所使用的,“保守置换”通常在蛋白质的一个或多个位点上交换一种氨基酸。这种取代可以是保守的。作为被视作保守置换的置换,具体而言,可以举出Ala向Ser或Thr的置换、Arg向Gln、His或Lys的置换、Asn向Glu、Gln、Lys、His或Asp的置换、Asp向Asn、Glu或Gln的置换、Cys向Ser或Ala的置换、Gln向Asn、Glu、Lys、His、Asp或Arg的置换、Glu向Gly、Asn、Gln、Lys或Asp的置换、Gly向Pro的置换、His向Asn、Lys、Gln、Arg或Tyr的置换、Ile向Leu、Met、Val或Phe的置换、Leu向Ile、Met、Val或Phe的置换、Lys向Asn、Glu、Gln、His或Arg的置换、Met向Ile、Leu、Val或Phe的置换、Phe向Trp、Tyr、Met、Ile或Leu的置换、Ser向Thr或Ala的置换、Thr向Ser或Ala的置换、Trp向Phe或Tyr的置换、Tyr向His、Phe或Trp的置换、及Val向Met、Ile或Leu的置换。此外,保守突变还包括起因于基因所来源的个体差异、株、种的差异等天然产生的突变。
本公开中的“序列同一性”和“同一性百分比”指两个或更多个多核苷酸或多肽之间相同(即同一)的核苷酸或氨基酸的百分比。两个或更多个多核苷酸或多肽之间的序列同一性可通过以下方法测定:将多核苷酸或多肽的核苷酸或氨基酸序列对准且对经对准的多核苷酸或多肽中含有相同核苷酸或氨基酸残基的位置数目进行评分,且将其与经对准的多核苷酸或多肽中含有不同核苷酸或氨基酸残基的位置数目进行比较。多核苷酸可例如通过含有不同核苷酸(即取代或突变)或缺失核苷酸(即一 个或两个多核苷酸中的核苷酸插入或核苷酸缺失)而在一个位置处不同。多肽可例如通过含有不同氨基酸(即取代或突变)或缺失氨基酸(即一个或两个多肽中的氨基酸插入或氨基酸缺失)而在一个位置处不同。序列同一性可通过用含有相同核苷酸或氨基酸残基的位置数目除以多核苷酸或多肽中氨基酸残基的总数来计算。举例而言,可通过用含有相同核苷酸或氨基酸残基的位置数目除以多核苷酸或多肽中核苷酸或氨基酸残基的总数且乘以100来计算同一性百分比。
示例性的,在本公开中,当使用序列比较算法或通过目视检查测量以最大的对应性进行比较和比对时,两个或多个序列或子序列具有至少40%、50%、60%、70%、80%、85%、90%、91%、92%、93%、94%、95%、96%、97%、98%或99%核苷酸或氨基酸残基的“序列同一性”或“同一性百分比”。“序列同一性”或“同一性百分比”的判断/计算可以基于序列任何合适的区域上。例如,长度至少约50个残基的区域、至少约100个残基的区域,至少约200个残基的区域,至少约400个残基的区域,或至少约500个残基的区域。在某些实施方案中,所述序列在任一或两个相比较的生物聚合物(就是核酸或多肽)的整个长度上基本相同。
如本公开所使用的,术语“反向互补序列”(Reverse Complementary Sequence)的含义为:和原始多核苷酸的序列的方向相反,并且与原始多核苷酸的序列也互补的序列。示例性的,如果原始多核苷酸序列为ACTGAAC,则其反向互补序列为GTTCAGT。
如本公开所使用的,术语“多核苷酸”指由核苷酸组成的聚合物。多核苷酸可以是单独片段的形式,也可以是更大的核苷酸序列结构的一个组成部分,其是从至少在数量或浓度上分离一次的核苷酸序列衍生而来的,能够通过标准分子生物学方法(例如,使用克隆载体)识别、操纵以及恢复序列及其组分核苷酸序列。当一个核苷酸序列通过一个DNA序列(即A、T、G、C)表示时,这也包括一个RNA序列(即A、U、G、C),其中“U”取代“T”。换句话说,“多核苷酸”指从其他核苷酸(单独的片段或整个片段)中去除的核苷酸聚合物,或者可以是一个较大核苷酸结构的组成部分或成分,如表达载体或多顺反子序列。多核苷酸包括DNA、RNA和cDNA序列。“重组多核苷酸”、“重组核酸分子”属于“多核苷酸”中的一种。
如本公开所使用的,术语“重组核酸分子”指具有在自然界中不连接在一起的序列的多核苷酸。重组多核苷酸可包括在合适的载体中,且该载体可用于转化至合适的宿主细胞。然后多核苷酸在重组宿主细胞中表达以产生例如“重组多肽”“重组蛋白”“融合蛋白”等。
如本公开所使用的,术语“连接肽”和“linker”可以替换使用,其能够将相同或者不同的多肽或者氨基酸进行连接。
连接肽包括柔性连接肽和刚性连接肽。在本公开的实施例中,连接肽选用柔性连接肽。其中,优选地,柔性连接肽选择:(Gly Gly Gly Gly Ser)
n,n=1~6之间的整数;或者(Gly Gly Gly Gly Thr)
n,n=1~6之间的整数。更优选的,本公开中的柔性连接肽选自(Gly Gly Gly Gly Ser)
2。
如本公开所使用的,术语“高严格条件”是指,对于长度为至少100个核苷酸的探针而言,遵循标准DNA印迹程序,在42℃处在5X SSPE(saline sodium phosphate EDTA)、0.3%SDS、200微克/ml剪切并变性的鲑精DNA和50%甲酰胺中预杂交和杂交12至24小时。最后在65℃处使用2X SSC、0.2%SDS将载体材料洗涤三次,每次15分钟。
如本公开所使用的,术语“非常高严格条件”是指,对于长度为至少100个核苷酸的探针而言,遵循标准DNA印迹程序,在42℃处在5X SSPE(saline sodium phosphate EDTA)、0.3%SDS、200微克/ml剪切并变性的鲑精DNA和50%甲酰胺中预杂交和杂交12至24小时。最后在70℃处使用2X SSC、0.2%SDS将载体材料洗涤三次,每次15分钟。
在本公开中,除非特别强调,否则术语“嵌合肽修饰的SIS膜”和“嵌合肽修饰的GBR膜”的含义相同,可以替换使用。
除非另外定义或由背景清楚指示,否则在本公开中的全部技术与科学术语具有如本公开所属领域的普通技术人员通常理解的相同含义。
技术方案
在本公开的技术方案中,说明书核苷酸和氨基酸序列表的编号所代表的含义如下所示:
SEQ ID NO:1示出了P1多肽(TKKTLRT+linker-1+Hst8)的氨基酸序列;
SEQ ID NO:2示出了P2多肽(TKKTLRT+linker-1+JH8195)的氨基酸序列;
SEQ ID NO:3示出了P3多肽(KELNLVY+linker-1+Hst8)的氨基酸序列;
SEQ ID NO:4示出了P4多肽(KELNLVY+linker-1+JH8195)的氨基酸序列;
SEQ ID NO:5示出了P5多肽(TKKTLRT+linker-1+Hst7)的氨基酸序列;
SEQ ID NO:6示出了P6多肽(TKKTLRT+linker-1+JH8944)的氨基酸序列;
SEQ ID NO:7示出了P7多肽(KELNLVY+linker-1+Hst7)的氨基酸序列;
SEQ ID NO:8示出了P8多肽(KELNLVY+linker-1+JH8944)的氨基酸序列;
SEQ ID NO:9示出了P9多肽(TKKTLRT+linker-1+Hst1)的氨基酸序列;
SEQ ID NO:10示出了P10多肽(TKKTLRT+linker-1+JH8194)的氨基酸序列;
SEQ ID NO:11示出了P11多肽(KELNLVY+linker-1+Hst1)的氨基酸序列;
SEQ ID NO:12示出了P12多肽(KELNLVY+linker-1+JH8194)的氨基酸序列;
SEQ ID NO:13示出了P13多肽(TKKTLRT+linker-2+Hst1)的氨基酸序列;
SEQ ID NO:14示出了P14多肽(TKKTLRT+linker-2+JH8194)的氨基酸序列;
SEQ ID NO:15示出了P15多肽(KELNLVY+linker-2+Hst1)的氨基酸序列;
SEQ ID NO:16示出了P16多肽(KELNLVY+linker-2+JH8194)的氨基酸序列。
除非另有强调,否则本公开的实施例中采用的以下通用实验方法(A)-(E)的步骤如下:
(A)制备SIS和嵌合肽修饰的SIS膜的方法:
SIS膜的制备方法如下:
(i)小肠粘膜下层材料的初处理
取小肠粘膜下层(Small Intestinal Submucosa,SIS)材料进行初处理。
在一个具体的实施方式中,本公开中的前述初处理包括对SIS材料进行分割、清洗和/或病毒灭活的步骤。
病毒灭活可采用低浓度过氧乙酸-乙醇溶液法灭活病毒,该步骤可在超声波清洗器中进行,其中过氧乙酸的体积百分含量可为0.05~0.2%,超声波振荡频率可为30~600rpm,超声波频率可为20~80KHZ,温度范围为4~40℃,然后在水或磷酸盐缓冲液中清洗。
(ii)免疫原去除处理
可以采用包括物理、化学和/或生物学方法去除经步骤(i)处理的小肠粘膜下层材料中的免疫原。包括冻融法、高低渗法、酸碱溶解法、去污剂剂法、酶法等方法进行去除。还可以多种方法相结合的方式进行免疫原的去除。优选超声辅助的方式,例如可在超声波清洗器中进行。首先将经步骤(i)处理的小肠粘膜下层材料置入清洗槽中,在清洗槽中注入脱细胞液,例如碱性溶液、盐溶液或含有酶的溶液,进行处理。将处理完成的材料进行清洗。
经步骤(ii)处理后的小肠粘膜下层材料可以作为嵌合肽修饰的SIS膜的膜材。
在另一个具体的实施方式中,还可以通过层叠前述经(ii)处理后的小肠粘膜下层材料的方式提高膜材的厚度和强度。
在另一个具体的实施方式中,为便于转运和处理还可以对前述层叠后的材料进行干燥处理。该干燥处理包括冻干和/或常温干燥处理。
嵌合肽修饰的SIS膜的制备方法如下:
将SIS膜切割成与不同培养板尺寸相匹配的圆形并且浸在低浓度(50μM)、中浓度(100μM)和高浓度(200μM)的嵌合肽溶液中10min。随后,将膜取出并且冷冻过夜,然后冻干以产生嵌合肽修饰的SIS膜(pSIS)。在后续的示例性实验中,用L-pSIS表示采用低浓度嵌合肽溶液浸泡的SIS膜,用M-pSIS表示采用中浓度嵌合肽溶液浸泡的SIS膜,用H-pSIS表示采用高浓度嵌合肽溶液浸泡的SIS膜。本处理过程中采用了50μM、100μM和200μM的嵌合肽溶液作为示意性对照,而并非对嵌合肽溶液浓度进行限制。
在另一个具体的实施方式中,还可以采用其它方式将嵌合肽修饰至SIS膜表面,例如将含有嵌合肽的溶液通过涂抹、喷涂或转印等方式施加至SIS膜表面,再进行干燥处理。
(B)细胞培养物的qRT-PCR分析方法:
通过使用Trizol(Gibco,USA)来提取总细胞RNA。使用GoScript
TM逆转录混合液(Promega,USA)来合成cDNA。使用GoTaq qPCR Master Mix(Promega,USA)在Roche LC480II系统(Roche,瑞士)上进行qRT-PCR分析。使用GAPDH作为内部对照通过
ΔΔCt方法来计算数据。引物的序列如表5中列出。
表5 Real-time PCR的引物序列
(C)细胞培养物的免疫印迹分析方法:
培养后,将蛋白通过电泳来分离并且转印至聚偏二氟乙烯(PVDF)膜(Sigma-Aldrich,USA)。在用5%牛血清白蛋白(BSA,Sigma‐Aldrich,USA)在室温下封闭1h后,在4℃下用1:1000一抗(Abcam,UK)探测PVDF膜过夜。然后,将它们用相应的二抗以1:5000的稀释度在室温下温育60min。并且使用Pierce
TMECL免疫印迹底物(Thermo Fisher Scientific,USA)来检测与抗体结合的蛋白。Image J软件用于定量分析。
(D)细胞培养物的免疫荧光染色方法:
将1×10
4个BMSC(Bone Mesenchymal Stem Cells)或OEC接种至24孔板上,用1mL SIS膜、M-pSIS膜或H-pSIS膜浸提液培养,然后用成骨诱导培养基培养BMSC。在相应的培养期后,将BMSC或OEC在4%甲醛中固定30min、用0.5%Triton X-100透化5min并且在5mg/mL BSA溶液中封闭1h。然后,将细胞用一抗(Abcam,UK)以1:200的稀释度在37℃下温育2h。此后,将它们用缀合有Cy3的抗兔/小鼠二抗(Abcam,UK)以1:200的稀释度温育1h,然后用1mg/mL DAPI(Solarbio,中国)染色10min。使用CLSM来进行可视化。
(E)统计学分析方法:
所有实验数据均表示为至少重复三次的平均值±标准偏差(SD)。使用带有Turkey’s检验的方差的单因素方差分析(ANOVA)来评估统计学分析。p值小于0.05被认为是具有显著的统计学差异(*p<0.05)。
实施例
本公开的其他目的、特征和优点将从以下详细描述中变得明显。但是,应当理解的是,详细描述和具体实施例(虽然表示本公开的具体实施方式)仅为解释性目的而给出,因为在阅读该详细说明后,在本公开的精神和范围内所作出的各种改变和修饰,对于本领域技术人员来说将变得显而易见。
本实施例中所用到的实验技术与实验方法,如无特殊说明均为常规技术方法,例如下列实施例中未注明具体条件的实验方法,通常按照常规条件如Sambrook等人,分子克隆:实验室手册(New York:Cold Spring Harbor Laboratory Press,1989)中所述的条件,或按照制造厂商所建议的条件。实施例中所使用的材料、试剂等,如无特殊说明,均可通过正规商业渠道获得。
实施例1:嵌合肽的设计和合成
为了验证嵌合肽的活性差异,本公开设计了包含多组不同功能的嵌合肽的组合。
通过Jill biochemical Shanghai Co.,Ltd,China公司合成本公开所涉及的嵌合多肽。
本公开中所提及的嵌合多肽的分组和具体序列如下。
Control-1:胶原结合肽(I型:TKKTLRT;III型:KELNLVY);柔性Linker:GGGGSGGGGS;功能肽:Hst8/JH8195。Control-1中的4条嵌合肽的序列如表1所示。
表1 Control-1嵌合肽序列
Control-2:胶原结合肽(I型:TKKTLRT;III型:KELNLVY);柔性Linker:GGGGSGGGGS;功能肽:Hst7/JH8944。Control-2中的4条嵌合肽的序列如表2所示。
表2 Control-2嵌合肽序列
pSIS-1:胶原结合肽(I型:TKKTLRT;III型:KELNLVY);柔性Linker:GGGGSGGGGS;功能肽:Hst1/JH8194;pSIS-1中的4条嵌合肽的序列如表3所示。其中,Hst1具有一定程度的愈合活性,JH8194具有一定程度的成骨活性和抗菌活性。
表3 pSIS-1嵌合肽序列
pSIS-2:胶原结合肽(I型:TKKTLRT;III型:KELNLVY);刚性Linker:EAAAKEAAAK;功能肽:Hst1/JH8194;pSIS-2中的4条嵌合肽的序列如表4所示。
表4 pSIS-2嵌合肽序列
实施例2:pSIS嵌合肽的结构预测
需要说明的是,实施例2中的pSIS对应于实施例1中的pSIS-1。
根据表3中的序列通过Fmoc固相肽合成法(Jill biochemical Shanghai Co.,Ltd,China)来合成嵌合肽至纯度为95%。将P9、P10用FITC标记并且将P11、P12用RB标记,用于CLSM。借助软件PSIPRED来分析二级结构。借助蛋白质分析软件Robetta来预测三级结构并且借助VMD来使其可视化。
通过PSIPRED和Robetta预测P9、P10、P11和P12的结构。嵌合肽由三部分组成:P9和P10:TKKTLRT(结合至SIS膜的I型胶原)、Hst1/JH8194(起到抗菌、成骨和愈合促进能力的功能)和GGGGSGGGGS(连接前两部分),P11和P12:KELNLVY(结合至SIS膜的III型胶原)、Hst1/JH8194和GGGGSGGGGS。
图1A示出了胶原嵌合肽P9和P10的设计详情和结构。图1B示出了胶原嵌合肽P11和P12的设计详情和结构。四种嵌合肽由胶原结合肽(TKKTLRT或KELNLVY)、功能性肽(Hst1或JH8194)和柔性接头(GGGGSGGGGS)构成。“GGGGS”是由甘氨酸(Gly)和丝氨酸(Ser)残基组成的最常用的柔性接头,其可确保连接功能性结构域的柔性(flexibility)和移动性(mobility)。在本公开中,“GGGGS”的拷贝数优选为2,以实现功能性结构域的适当的分离并避免相互干扰。
图1A和图1B的结果显示,P9-P12均具有一种或多种基本结构,例如α螺旋、β折叠和无规卷曲,并且它们的空间结构不同。
实施例3:嵌合肽修饰的SIS膜(pSIS)的制备、形态学观察和生物相容性
需要说明的是,实施例3中的pSIS对应于实施例1中的pSIS-1。
将SIS膜切割成与不同培养板尺寸相匹配的圆形并且浸在低浓度(50μM)、中浓度(100μM)和高浓度(200μM)的嵌合肽溶液中10min。随后,将膜取出并且冷冻过夜,然后冻干以产生嵌合肽修饰的SIS膜(pSIS)。将样品标记为L-pSIS组、M-pSIS组和H-pSIS组。在用金溅镀之后,通过SEM(Gemini 300,Zeiss,德国)来观察SIS膜和pSIS膜的表面形态。
此外,将在1000μL含有10%胎牛血清(FBS,Gibco,USA)的Dulbecco改良的Eagle培养基(DMEM,Gibco,USA)中的1×10
4个BMSC或OEC接种至24孔板上并且在37℃、5%CO
2下培养。在1、2、4和6天后,将100μL CCK-8溶液(Solarbio,China)添加至各样品。在37℃、5%CO
2下温育4h后,通过测量在450nm处的吸光度(OD)来确定相对细胞活力。
图2A示出了pSIS膜的示意图。图2B示出了CLSM观察的嵌合肽的结合。其中,P9和P10由罗丹明B(RB)标记,P11和P12由异硫氰酸荧光素(FITC)标记。图2C示出了通过SEM观察冻干SIS和pSIS的表面形态。其中,不同表示的含义如下:SIS:浸在PBS中;L-pSIS:浸在50μM嵌合肽溶液中;M-pSIS:浸在100μM嵌合肽溶液中;H-pSIS:浸在200μM嵌合肽溶液中)。图2D示出了通过CCK-8的BMSC和OEC的增殖能力。
其后,将SIS膜浸在50μM(L-pSIS)、100μM(M-pSIS)和200μM(H-pSIS)嵌合肽溶液中以观察嵌合肽在SIS膜的表面上的结合,我们使用共聚焦激光扫描显微镜(CLSM)和扫描电子显微镜(SEM)来检测。随着浓度增加,SIS膜上的荧光标记增加(图2B),表明更多的嵌合肽结合。特别地,在用200μM嵌合肽溶液浸泡后,SIS膜(H-pSIS)的超过85%的荧光面积实现有效结合。然后,通过SEM观察冻干后的pSIS的表面形态(图2C)。与SIS相比,pSIS仍维持相似的结构并具有通过大量嵌合肽与SIS膜的特异性组合所形成的小颗粒样物质。冻干处理不影响SIS膜的结构和嵌合肽的稳定结合。嵌合肽溶液的浓度越高,pSIS的表面上的颗粒越多。然而,由于嵌合肽的极小分子量,SEM仅可观察大量的聚集颗粒,这与CLSM下观察到的结合率可能不匹配。
生物相容性,材料和宿主之间的相互作用,是材料的基本特征,以确保应用期间患者的安全。通过CCK-8研究pSIS对细胞增殖随时间的作用。其显示BMSC和OEC在pSIS的表面上对数增殖(图2D)。在第二天,BMSC在H-pSIS膜上的增殖能力显著低于其他组,但随着时间延长,增殖能力提高并与其他组无统计学差异。发现pSIS对BMSC或OEC的增殖无显著作用,为它们的进一步应用提供保证。因此,考虑到CLSM、SEM和CCK-8的结果,我们选择具有较高嵌合肽的结合率的M-pSIS 和H-pSIS用于后续实验中的进一步研究。
实施例4:pSIS的体外抗菌活性
需要说明的是,实施例4中的pSIS对应于实施例1中的pSIS-1。
将血链球菌(ATCC 10556)和格氏链球菌(ATCC 51656)在脑心浸液(BHI)琼脂平板中在37℃下培养18h。然后,将细菌以1×10
7CFU/ml(CFU,菌落形成单位)重悬,作为主要接种物。在100mm细菌培养皿中,接种混合有100μl细菌溶液的20ml BHI溶液。凝固后,将SIS膜、M-pSIS膜和H-pSIS膜置于中心,并且使用空白孔作为对照。24h后,观察抑菌圈并拍照。
此外,在将主要接种物接种在SIS膜、M-pSIS膜和H-pSIS膜上并且温育24h后,将膜用PBS洗涤三次,然后在新板上用活/死染色试剂盒(Life Technologies Corporation,CA)染色。可以通过CLSM来观察死细菌、活细菌和Alexa Fluor 405标记的肽。
图3A示出了血链球菌(S.sanguis)和格氏链球菌(S.gordonii)的抑菌圈(左上:空白对照;右上:SIS;左下:M-pSIS;右下:H-pSIS)。图3B示出了在空白孔、SIS、M-pSIS或H-pSIS中培养细菌24小时后染色的CLSM图像。
为了调查pSIS针对血链球菌(S.sanguis)和格氏链球菌(S.gordonii)的抗菌活性,检测抑菌圈(inhibitory ring)和CLSM(图3A-图3B)。尽管SIS可产生微小的抑菌圈,但结果是统计学不显著的,同时M-pSIS和H-pSIS可对两种细菌产生明显的抑菌圈(图3A)。此外,CLSM图像显示细菌的活/死菌落(图3B)。与空白孔相比,SIS组具有少量的死细菌。相反地,随着肽浓度的增加,M-pSIS和H-pSIS的死细菌依次增加,表明嵌合肽增强SIS膜的抗菌活性,并且H-pSIS是最强的。
菌斑生物膜是多物种微生物群落,其可增强细菌对宿主防御系统和抗菌剂的抵抗力,最终导致炎症。格氏链球菌和血链球菌是牙菌斑生物膜的早期定植细菌。上述菌群定植的及时和早期控制将有利于防止GBR手术后的口腔感染和炎症。JH8194是口腔抗菌肽Hst5的衍生肽。AMP及其衍生物可通过“离子交换机制”在细菌膜上交换二价阳离子(例如,Mg
2+和Ca
2+)以破坏细胞质膜的稳定性,导致细胞死亡。此外,带正电的AMP可通过静电相互作用吸附在带负电的细胞质膜上,并插入磷脂双分子层以在膜上形成孔或甚至更大的缺陷,最终导致细胞质泄漏和细菌死亡。
实施例5:pSIS对BMSC的成骨相关因子表达的作用
需要说明的是,实施例5中的pSIS对应于实施例1中的pSIS-1。
pSIS的体外细胞迁移活性的测定方法:
将SIS膜、M-pSIS膜或H-pSIS膜和500μL培养基(2%FBS)置于transwell板的下部隔室中,并且仅具有培养基的空白孔用作对照。然后,将在500μL培养基(2%FBS)中的1×10
4个OEC接种至transwell插入皿上(Thermo Fisher Scientific,USA)。24h后,小心地除去transwell插入皿内的培养基。将细胞用4%多聚甲醛固定30min,用0.01%Triton X‐100(Sigma‐Aldrich,USA)透化5min并且用1%结晶紫(Sigma‐Aldrich,USA)染色。通过用棉签轻轻擦拭来除去未迁移的细胞。然后,用倒置荧光显微镜(Olympus IX71,日本)观察迁移的细胞并且使其成像。
将1×10
5个OEC接种至具有无菌的SIS膜、M-pSIS膜和H-pSIS膜的6孔板上并且培养24h。然后,借助细胞刮棒将细胞刮下并且通过1000rpm离心3min来收集细胞。每个样品需要1×10
6个OEC。随后,由RNA-seq(Beijing Nuohe Zhiyuan biological Mdt InfoTech Ltd,中国)商业检测差异表达的基因。在将OEC培养24h后,使用定量实时聚合酶链反应(qRT-PCR)分析、免疫印迹分析和免疫荧光染色来检测差异表达的因子。
pSIS的体外成骨活性的测定方法如下:
该工作中的动物实验由天津医科大学的学术医学中心的动物伦理委员会批准。简言之,将实验分为空白对照组、SIS组、Bio-Gide组和H-pSIS组。使Sprague-Dawley(SD)大鼠(250~280g,雄性,6-8周)通过吸入异氟烷而被麻醉。然后,通过切开皮肤来暴露顶颅盖(Parietal Calvarium)。随后,用环锯在颅骨中央形成直径为8mm的全层缺损(Full-Thickness Defect)。对空白对照组不进行处理,其它三组将相应的膜置于缺损上。最后,将伤口缝合。
在12周结束时,将大鼠安乐死。将骨缺损取出并且在4%多聚甲醛中固定24h。然后,将样品 用Micro-CT(SkyScan 1276,德国)来扫描,以进行标准化重建和评价新骨形成。接下来,将样品用快速脱钙液(Rapid Cal Immuno,ZS-Bio,中国)脱钙7d。在脱水和用石蜡包埋后,将样品以5μm的厚度切片。将H&E和Masson三色染色(Solarbio,中国)用于组织学分析。将稀释度为1:200的一抗抗-OCN、抗-COL1和荧光缀合的二抗IgG(Abcam,UK)用于免疫组织化学法。使用组织切片的全景成像的定量分析系统(Quantitative Analysis System for Whole Landscape Imaging of Tissue Slices)(Vectra Polaris,PerkinElmer,USA)来观察切片。
制备含有10mMβ-甘油磷酸酯(Sigma,USA)、100nM地塞米松(Sigma,USA)和50μM抗坏血酸盐(Sigma,USA)的成骨诱导培养基。将1×10
5个BMSC接种在6孔板中的无菌SIS膜、M-pSIS膜和H-pSIS膜上,用培养基培养3天,然后使用成骨诱导培养基替换。7天、14天和21天后,检测BMP2、RUNX2、ALP和OPN的表达。
图4A示出了成骨相关基因的qPCR分析结果(*表示p<0.05)。图4B示出了成骨相关蛋白的免疫印迹分析结果。图4C示出了免疫印迹分析的定量分析结果(*表示p<0.05)。图4D示出了BMSC中成骨相关蛋白定位的免疫荧光分析结果(BMP2、RUNX2、ALP、OPN和细胞核)。
成骨作用是GBR技术的关键。还评价pSIS对BMSC的成骨相关因子的表达的作用(图4A-图4D)。骨形态发生蛋白-2(BMP2)、Runt相关转录因子2(RUNX2)、碱性磷酸酶(ALP)和骨桥蛋白(OPN)是经典成骨因子。如图4A中所示,pSIS组的上述成骨因子的mRNA水平显著高于空白对照和SIS。类似地,免疫印迹分析也确证了该趋势(图4B和图4C),表明嵌合肽修饰的SIS膜可进一步促进成骨相关因子的表达。然后,通过CLSM确定BMP2、RUNX2、ALP和OPN在细胞中的定位(图4D)。可以看到,M-pSIS和H-pSIS的红色荧光显著高于SIS的,并且H-pSIS表达最高的荧光信号,这也确证了上述趋势。蛋白质分布在远离细胞膜的细胞质中。上述结果表明JH8194具有一定的成骨功能。
实施例6:pSIS对OEC迁移的作用和可能的信号通路
需要说明的是,实施例6中的pSIS对应于实施例1中的pSIS-1。
pSIS的体内促愈合能力的检测方法如下:
使SD大鼠通过吸入异氟烷而被麻醉并且将其固定在俯卧位。在除去背部的毛发后,在脊柱的两侧通过使用钻孔机(直径为2cm)形成四个对称的圆形标记。沿着圆形标记,用解剖刀切下整个皮肤并且将其分为如下几组:空白对照(Blank)、SIS膜组、Bio-Gide膜组和H-pSIS膜组,并且将膜缝合和用皮下组织固定。每只大鼠分开饲养以避免其互相抓咬。H&E、Masson三色染色和免疫组织化学法的具体方法与前述相同。通过免疫组织化学法来检测ITG-β1和ITG-α3的表达。
图5A示出了Transwell实验检测细胞迁移能力的结果。图5B示出了迁移细胞的分析的结果(*表示p<0.05)。图5C示出了RNA-seq筛选的差异表达基因的结果。图5D示出了不同基因的维恩图(Venn diagram)。图5E示出了迁移相关基因的qPCR分析的结果。图5F示出了迁移相关蛋白的蛋白质印迹分析的结果。图5G示出了蛋白质印迹分析的定量分析的结果(*表示p<0.05)。图5H示出了OEC中迁移相关蛋白定位的免疫荧光分析的结果(ITG-α3、ITG-β1和细胞核)。
GBR中的伤口愈合过程涉及细胞增殖和迁移、伤口收缩和血管发生、胶原沉积和重塑。由多个步骤的过程构成的细胞迁移是愈合修复所必需的。Hst1的序列"SHREFPFYGDYGS"含有促进细胞迁移所必需的最小元素,其可以促进口腔角质形成细胞、口腔上皮细胞和牙龈成纤维细胞的迁移。通过Transwell实验检测pSIS对OEC的迁移的作用(图5A和图5B)。SIS和pSIS可以促进OEC的迁移,其中H-pSIS具有最强能力,表明嵌合肽修饰的SIS膜可以进一步促进细胞迁移,并且会有助于软组织的愈合。
随后,为了探索嵌合肽促进OEC迁移的具体机制,使用一种新型的高通量测序方法RNA-seq分析在SIS、M-pSIS和H-pSIS上培养24h的OEC的差异表达基因(图5C)。与SIS组相比,M-pSIS组中790个基因上调,833个基因下调,H-pSIS组中756个基因上调,735个基因下调。维恩图显示844个基因具有相同的趋势。通过对不同基因的聚类分析,我们发现ITG-β1与细胞迁移有关(图5D)。
然而,大多数整联蛋白是由非共价连接的亚基α和β形成的异二聚体分子,它介导细胞的粘附 和迁移并在伤口修复中起重要作用。整联蛋白α3β1与细胞迁移和伤口愈合密切相关。它可以通过加速角质形成细胞的迁移来促进再上皮化,以协助上皮伤口的愈合。在GBR区域,整联蛋白α3β1可以与未加工的层粘连蛋白5结合,然后介导相连的上皮细胞的迁移。因此,检测OEC的ITG-α3和ITG-β1的表达。H-pSIS组中ITG-α3和ITG-β1的表达高于SIS组(图5E-图5G),表明嵌合肽可以促进迁移相关因子的表达。然后,通过CLSM确定细胞中ITG-α3和ITG-β1的定位(图5H)。可以看出,H-pSIS组的前述蛋白的荧光强于SIS组,并且蛋白质分布在远离细胞膜的细胞质中。
实施例7:体内pSIS成骨能力的评价
需要说明的是,实施例7中的pSIS对应于实施例1中的pSIS-1。
图6A示出了用SIS、Bio-Gide和H-pSIS膜覆盖的骨缺损的三维重建和矢状图像。图6B示出了不同组的骨缺损的BV/TV结果(*表示p<0.05)。图6C示出了H&E染色结果。图6D示出了Masson的三色染色的结果。图6E示出了OCN和COL1表达的免疫组织化学分析结果。
在动物实验中,我们选择效果最好的H-pSIS膜用于进一步的体内研究(图6A-图6E)。在大鼠中制作了无法自我修复的8mm圆形缺损的临界尺寸缺损(CSD)模型,以评价骨再生。此外,将为改善GBR中任何来源的骨缺损的骨化而特别开发的Bio-Gide膜(商业购买)用作对照。如图6A所示,所有组的微计算机断层扫描(CT)图像均显示,新骨的形成是从缺损边缘向中心发展的。术后十二周,SIS组和Bio-Gide组的BV/TV高于空白对照,但SIS和Bio-Gide之间无显著差异。此外,H-pSIS组显示出在体内极大改善的成骨作用(图6A-图6B)。
通过苏木精-曙红(H&E)和Masson三色染色进行组织学分析,以评价胶原和新骨组织的生长以及淋巴细胞的浸润。胶原是骨骼的重要组成部分。包括I型胶原在内的成骨细胞分泌的ECM可从最初的无定形和非结晶性转变为更多的结晶性,从而逐渐导致成骨作用。此外,矿化是成骨细胞促进骨形成的主要过程。胶原作为模板,还可以引发和传播矿化作用。因此,胶原含量与骨形成密切相关。如图6C-图6D所示,空白对照组的缺损区域主要由纤维组织构成,胶原较少,没有明显的新骨形成迹象。然而,SIS组、Bio-Gide组和H-pSIS组的骨缺损区域富含胶原,并且新骨在残端(stump)周围形成,有少量甚至无纤维结缔组织浸润。同时,所有组的淋巴细胞浸润均低,表明没有炎症反应(图6C)。通过免疫组化法检测骨钙素(OCN)和I型胶原(COL1)的表达(图6E)。术后十二周,对照组未见明显的OCN和COLI染色,而在SIS、Bio-Gide和H-pSIS组中高表达,H-pSIS组最高。
以上结果表明,H-pSIS可以有效地防止纤维结缔组织长入缺损区域,为骨形成提供空间。pSIS优异的骨再生能力有助于解决由牙周炎或牙齿缺失引起的牙槽骨缺失以及牙种植体区域骨质不足的问题。此外,其独特的抗感染能力有助于抵抗和预防组织炎症和感染,从而修复感染性骨缺损,有望解决修复与感染相关的骨缺损的临床难题。
实施例8:体内pSIS促进愈合能力的评价
需要说明的是,实施例8中的pSIS对应于实施例1中的pSIS-1。
图7A示出了H&E染色结果(线段:非上皮伤口的长度)。图7B示出了新上皮化长度(*表示p<0.05)。图7C示出了Masson的三色染色结果(线段:伤口的非胶原纤维的长度)。图7D示出了胶原纤维长度(*表示p<0.05)。图7E示出了ITG-α3和ITG-β1表达的免疫组织化学分析结果。
为了评价软组织缺损区域的再上皮化和胶原蛋白含量,通过H&E和Masson三色染色进行了组织学分析(图7A-图7D)。如图7A和图7B所示,实验用大鼠在手术后1和2周,SIS组和H-pSIS组的再上皮化程度显著高于Bio-Gide和空白对照,H-pSIS最高。再上皮化是伤口愈合的基本特征,这与角质形成细胞的定向迁移有关。pSIS膜可通过促进细胞迁移来促进再上皮化,这有助于伤口的快速愈合。与骨形成一样,治愈伤口的成功需要局部合成大量胶原,H-pSIS的软组织缺损区域的胶原含量明显高于其他组(图7C-图7D),表明H-pSIS具有更好的促进软组织愈合的能力。通过免疫组织化学检测ITG-α3和ITG-β1的表达,阳性区域描述在图7E中。SIS、Bio-Gide和H-pSIS组中ITG-α3和ITG-β1的水平高于对照组,而H-pSIS组最高。
与此同时,前述大鼠实验在手术后1和2周后的愈合结果如图8所示,发现H-pSIS组具有更好的愈合效果。
这些结果表明,嵌合肽修饰的SIS膜促进上皮细胞迁移和胶原沉积,这与ITG-α3和ITG-β1的表达增加有关。
实施例9:不同嵌合肽修饰的SIS膜的成骨效果、抑制炎症效果和愈合效果比较
采用本公开中实施例5-8中所记载的方法,利用本公开中合成的不同的嵌合肽(P1-P8,P13-P16)修饰的SIS膜的成骨效果、抑制炎症效果和愈合效果进行比较。
成骨效果的比较结果如图9所示;抑制炎症效果如图10所示;愈合效果如图11所示。
图9-图11中,Control-1为P1-P4所示的嵌合肽的混合;Control-2为P5-P8所示的嵌合肽的混合;pSIS-1为P9-P12所示的嵌合肽的混合;pSIS-2为P13-P16所示的嵌合肽的混合。
从图9-图11的实验结果来看,pSIS-1具有更好的成骨效果、抑制炎症效果和愈合效果。
简而言之,GBR后促进伤口的早期愈合是预防微生物感染和生物材料暴露并发症的有效方法。目前,某些GBR膜在引导软组织愈合方面具有一定作用,但性能单一且作用有限。本公开开发的嵌合肽修饰的SIS膜可同时发挥抗菌作用,促进软组织愈合和骨再生,极大地丰富了GBR膜的性能,本公开证明了嵌合肽修饰的SIS膜(即GBR膜)可以用于感染性骨缺损的临床治疗。
本公开的上述实施例仅是为清楚地说明本公开所作的举例,而并非是对本公开的实施方式的限定。对于所属领域的普通技术人员来说,在上述说明的基础上还可以做出其它不同形式的变化或变动。这里无需也无法对所有的实施方式予以穷举。凡在本公开的精神和原则之内所作的任何修改、等同替换和改进等,均应包含在本公开权利要求的保护范围之内。
Claims (10)
- 一种嵌合肽修饰的SIS膜,其中,所述嵌合肽的序列包含由如下序列所示的序列组成的组中的至少一种:(i)如SEQ ID NO:9所示的序列、如SEQ ID NO:10所示的序列、如SEQ ID NO:11所示的序列、如SEQ ID NO:12所示的序列组成的组;(ii)和(i)所示的序列相比,存在保守置换的序列。
- 根据权利要求1所述的嵌合肽修饰的SIS膜,其中,所述嵌合肽的序列由如下序列组成的组中的至少一种组成:(i)如SEQ ID NO:9所示的序列、如SEQ ID NO:10所示的序列、如SEQ ID NO:11所示的序列、如SEQ ID NO:12所示的序列组成的组;(ii)和(i)所示的序列相比,存在保守置换的序列。
- 根据权利要求1-2任一项所述的嵌合肽修饰的SIS膜,其中,所述修饰的方法包括以下步骤:(a)将所述嵌合肽溶解至溶剂中,得到含有嵌合肽的溶解液;(b)将步骤(a)得到的溶解液施加至所述SIS膜的表面;优选的,将所述SIS膜浸入步骤(a)得到的溶解液中;(c)干燥表面带有所述溶解液的所述SIS膜。
- 根据权利要求1-3任一项所述的嵌合肽修饰的SIS膜,其中,所述SIS膜的制备方法包括以下步骤:(i)取小肠粘膜下层材料进行初处理;(ii)将经步骤(i)得到的小肠粘膜下层材料进行免疫原去除处理。
- 根据权利要求4所述的嵌合肽修饰的SIS膜,其中,所述SIS膜的制备方法还包括以下步骤:(iii)将经步骤(ii)得到的小肠粘膜下层材料进行层叠;(iv)将层叠后的小肠粘膜下层材料进行干燥处理。
- 一种嵌合肽修饰的SIS膜的制备方法,其中,所述方法包括以下步骤:(a)将所述嵌合肽溶解至溶剂中,得到含有嵌合肽的溶解液;(b)将步骤(a)得到的溶解液施加至所述SIS膜表面;优选的,将所述SIS膜浸入步骤(a)得到的溶解液中;(c)干燥表面带有所述溶解液的所述SIS膜,得到所述嵌合肽修饰的SIS膜;其中,所述嵌合肽的序列包含由如下序列所示的序列组成的组中的至少一种,或者所述嵌合肽的序列由如下序列组成的组中的至少一种组成:(i)如SEQ ID NO:9所示的序列、如SEQ ID NO:10所示的序列、如SEQ ID NO:11所示的序列、如SEQ ID NO:12所示的序列组成的组;(ii)和(i)所示的序列相比,存在保守置换的序列。
- 根据权利要求6所述的嵌合肽修饰的SIS膜的制备方法,其中,所述SIS膜的制备方法包括以下步骤:(i)取小肠粘膜下层材料进行初处理;(ii)将经步骤(i)得到的小肠粘膜下层材料进行免疫原去除处理。
- 根据权利要求7所述的嵌合肽修饰的SIS膜的制备方法,其中,所述SIS膜的制备方法还包括以下步骤:(iii)将经步骤(ii)得到的小肠粘膜下层材料进行层叠;(iv)将层叠后的小肠粘膜下层材料进行干燥处理。
- 根据权利要求1-5任一项所述的嵌合肽修饰的SIS膜或根据权利要求6-8任一项所述的嵌合肽修饰的SIS膜的制备方法得到的嵌合肽修饰的SIS膜在如下(a)-(d)至少一种中的用途:(a)作为或制备抗菌的生物材料;(b)作为或制备成骨的生物材料;(c)作为或制备促进愈合的生物材料;(d)作为或制备治疗感染性骨缺损的生物材料。
- 一种治疗感染性骨缺损的方法,其中,所述方法包括向受试者施用如权利要求1-5任一项所述的嵌合肽修饰的SIS膜或根据权利要求6-8任一项所述的嵌合肽修饰的SIS膜的制备方法得到的嵌合肽修饰的SIS膜的步骤。
Priority Applications (2)
| Application Number | Priority Date | Filing Date | Title |
|---|---|---|---|
| EP21938694.3A EP4331631A1 (en) | 2021-04-27 | 2021-06-02 | Chimeric peptide-modified sis membrane, preparation method therefor and application thereof |
| CN202180001723.1A CN113490517B (zh) | 2021-04-27 | 2021-06-02 | 一种嵌合肽修饰的sis膜、其制备方法及应用 |
Applications Claiming Priority (2)
| Application Number | Priority Date | Filing Date | Title |
|---|---|---|---|
| CN202110459709 | 2021-04-27 | ||
| CN202110459709.8 | 2021-04-27 |
Publications (1)
| Publication Number | Publication Date |
|---|---|
| WO2022227225A1 true WO2022227225A1 (zh) | 2022-11-03 |
Family
ID=83847673
Family Applications (1)
| Application Number | Title | Priority Date | Filing Date |
|---|---|---|---|
| PCT/CN2021/097851 WO2022227225A1 (zh) | 2021-04-27 | 2021-06-02 | 一种嵌合肽修饰的sis膜、其制备方法及应用 |
Country Status (1)
| Country | Link |
|---|---|
| WO (1) | WO2022227225A1 (zh) |
Citations (3)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| US5912230A (en) * | 1991-11-01 | 1999-06-15 | Periodontix, Inc. | Anti-fungal and anti-bacterial histatin-based peptides |
| WO2019075213A1 (en) * | 2017-10-11 | 2019-04-18 | Northwestern University | HEPARIN CONJUGATED TO PEPTIDES BINDING TO COLLAGEN FOR TARGETING BIOLOGICAL AND SYNTHETIC FABRICS |
| CN111050778A (zh) * | 2017-07-07 | 2020-04-21 | 斯米克Ip有限公司 | 具有化学修饰的骨架的生物缀合物 |
-
2021
- 2021-06-02 WO PCT/CN2021/097851 patent/WO2022227225A1/zh active Application Filing
Patent Citations (3)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| US5912230A (en) * | 1991-11-01 | 1999-06-15 | Periodontix, Inc. | Anti-fungal and anti-bacterial histatin-based peptides |
| CN111050778A (zh) * | 2017-07-07 | 2020-04-21 | 斯米克Ip有限公司 | 具有化学修饰的骨架的生物缀合物 |
| WO2019075213A1 (en) * | 2017-10-11 | 2019-04-18 | Northwestern University | HEPARIN CONJUGATED TO PEPTIDES BINDING TO COLLAGEN FOR TARGETING BIOLOGICAL AND SYNTHETIC FABRICS |
Non-Patent Citations (5)
| Title |
|---|
| LOZEAU, L. D. ET AL.: "Collagen tethering of synthetic human antimicrobial peptides cathelicidin LL37 and its effects on antimicrobial activity and cytotoxicity", ACTA BIOMATERIALIA, vol. 52, 23 December 2016 (2016-12-23), XP029968294, DOI: 10.1016/j.actbio.2016.12.047 * |
| MU YUZHU, SHIQING MA,PENGFEI WEI,YONGLAN WANG,WEI JING,YIFAN ZHAO,LEI ZHANG,JINZHE WU,BO ZHAO,JIAYIN DENG,ZIHAO LIU: "Multifunctional Modification of SIS Membrane with Chimeric Peptides to Promote Its Antibacterial, Osteogenic, and Healing-Promoting Abilities for Applying to GBR", ADV. FUNCT. MATER., vol. 31, no. 31, 28 May 2021 (2021-05-28), pages e2101452, XP055982062, DOI: 10.1002/adfm.202101452 * |
| SAMBROOK ET AL.: "Molecular Cloning: A Laboratory Manual (New York", 1989, COLD SPRING HARBOR LABORATORY PRESS |
| SONIA MELINO, CELESTE SANTONE, PAOLO DI NARDO, BIBUDHENDRA SARKAR: "Histatins: salivary peptides with copper(II)- and zinc(II)-binding motifs : Perspectives for biomedical applications", THE FEBS JOURNAL, WILEY-BLACKWELL PUBLISHING LTD., GB, vol. 281, no. 3, 1 February 2014 (2014-02-01), GB , pages 657 - 672, XP055594609, ISSN: 1742-464X, DOI: 10.1111/febs.12612 * |
| YANG KAI, YUMIN ZHANG, NAILI ZHANG, WEIJUN XU, LI BAOXING: "RECENT PROGRESS OF SMALL INTESTINAL SUBMUCOSA IN APPLICATION RESEARCH OF TISSUE REPAIR AND RECONSTRUCTION", CHINESE JOURNAL OF REPARATIVE AND RECONSTRUCTIVE SURGERY, vol. 27, no. 9, 15 August 2013 (2013-08-15), pages 1138 - 1143, XP055982058, DOI: 10.7507/1002-1892.20130248 * |
Similar Documents
| Publication | Publication Date | Title |
|---|---|---|
| CN113490517B (zh) | 一种嵌合肽修饰的sis膜、其制备方法及应用 | |
| US10449271B2 (en) | Matrix scaffold with antimicrobial activity | |
| US8846860B2 (en) | Methods of decorating hydroxyapatite biomaterials with modular biologically active molecules | |
| Deutzmann et al. | Molecular, biochemical and functional analysis of a novel and developmentally important fibrillar collagen (Hcol-I) in hydra | |
| Mu et al. | Multifunctional modification of SIS membrane with chimeric peptides to promote its antibacterial, osteogenic, and healing‐promoting abilities for applying to GBR | |
| JP2002542823A (ja) | ラミニン2とその使用方法 | |
| CA2965770C (en) | Treatment of cornea using laminin | |
| CA2438334C (en) | Ocular tear growth factor-like protein | |
| WO2022227225A1 (zh) | 一种嵌合肽修饰的sis膜、其制备方法及应用 | |
| Liu et al. | Functionalized self-assembled peptide RAD/Dentonin hydrogel scaffold promotes dental pulp regeneration | |
| JP3795805B2 (ja) | 細胞接着および創傷治癒のための方法 | |
| Cheng et al. | Hydroxyapatite‐Coated Small Intestinal Submucosa Membranes Enhanced Periodontal Tissue Regeneration through Immunomodulation and Osteogenesis via BMP‐2/Smad Signaling Pathway | |
| US20240041987A1 (en) | Novel bioactive peptide combinations and uses thereof | |
| WO2006041205A1 (ja) | 血管形成促進剤 | |
| EP4406563A1 (en) | Modified sis membrane for tissue repair, preparation method therefor and use thereof | |
| WO2023246135A1 (zh) | 肽、肽修饰的sis膜、其制备方法及应用 | |
| WO2011077086A2 (en) | Agents having tissue generative activity | |
| CN116271241B (zh) | 用于组织修复的改性非对称sis膜、其制备方法及应用 | |
| WO2023134090A1 (en) | Use of a neuropeptide in treating bone-injury disease | |
| CN112245567A (zh) | 一种抗菌及促成骨复合支架材料 | |
| JP5346145B2 (ja) | 神経再生用組成物 | |
| CN115926003B (zh) | 一种超分子弹性蛋白及其制备的水凝胶和应用 | |
| Seon et al. | Peptide derived from stromal cell-derived factor 1δ enhances the in vitro expression of osteogenic proteins via bone marrow stromal cell differentiation and promotes bone formation in in vivo models | |
| CN117659208A (zh) | 一种融合肽、包含其的复合支架、制备方法和用途 | |
| CN115835876A (zh) | 神经肽在治疗骨损伤疾病中的用途 |
Legal Events
| Date | Code | Title | Description |
|---|---|---|---|
| 121 | Ep: the epo has been informed by wipo that ep was designated in this application |
Ref document number: 21938694 Country of ref document: EP Kind code of ref document: A1 |
|
| WWE | Wipo information: entry into national phase |
Ref document number: 2021938694 Country of ref document: EP |
|
| NENP | Non-entry into the national phase |
Ref country code: DE |
|
| ENP | Entry into the national phase |
Ref document number: 2021938694 Country of ref document: EP Effective date: 20231127 |