WO2022068919A1 - 结合pd-1抗体的肽及其应用 - Google Patents
结合pd-1抗体的肽及其应用 Download PDFInfo
- Publication number
- WO2022068919A1 WO2022068919A1 PCT/CN2021/122062 CN2021122062W WO2022068919A1 WO 2022068919 A1 WO2022068919 A1 WO 2022068919A1 CN 2021122062 W CN2021122062 W CN 2021122062W WO 2022068919 A1 WO2022068919 A1 WO 2022068919A1
- Authority
- WO
- WIPO (PCT)
- Prior art keywords
- seq
- amino acids
- protein
- fab
- positions
- Prior art date
Links
- 230000027455 binding Effects 0.000 title claims abstract description 38
- 108090000765 processed proteins & peptides Proteins 0.000 title claims abstract description 24
- 102100040678 Programmed cell death protein 1 Human genes 0.000 claims abstract description 59
- 101710089372 Programmed cell death protein 1 Proteins 0.000 claims abstract description 57
- 239000012634 fragment Substances 0.000 claims abstract description 21
- 239000000427 antigen Substances 0.000 claims abstract description 16
- 102000036639 antigens Human genes 0.000 claims abstract description 16
- 108091007433 antigens Proteins 0.000 claims abstract description 16
- 125000003275 alpha amino acid group Chemical group 0.000 claims abstract 6
- 150000001413 amino acids Chemical group 0.000 claims description 63
- 108090000623 proteins and genes Proteins 0.000 claims description 11
- SHZGCJCMOBCMKK-UHFFFAOYSA-N D-mannomethylose Natural products CC1OC(O)C(O)C(O)C1O SHZGCJCMOBCMKK-UHFFFAOYSA-N 0.000 claims description 8
- SHZGCJCMOBCMKK-DHVFOXMCSA-N L-fucopyranose Chemical compound C[C@@H]1OC(O)[C@@H](O)[C@H](O)[C@@H]1O SHZGCJCMOBCMKK-DHVFOXMCSA-N 0.000 claims description 8
- 102000004169 proteins and genes Human genes 0.000 claims description 8
- PNNNRSAQSRJVSB-SLPGGIOYSA-N Fucose Natural products C[C@H](O)[C@@H](O)[C@H](O)[C@H](O)C=O PNNNRSAQSRJVSB-SLPGGIOYSA-N 0.000 claims description 6
- 108010054477 Immunoglobulin Fab Fragments Proteins 0.000 claims description 6
- 102000001706 Immunoglobulin Fab Fragments Human genes 0.000 claims description 6
- WQZGKKKJIJFFOK-QTVWNMPRSA-N D-mannopyranose Chemical compound OC[C@H]1OC(O)[C@@H](O)[C@@H](O)[C@@H]1O WQZGKKKJIJFFOK-QTVWNMPRSA-N 0.000 claims description 5
- OVRNDRQMDRJTHS-FMDGEEDCSA-N N-acetyl-beta-D-glucosamine Chemical compound CC(=O)N[C@H]1[C@H](O)O[C@H](CO)[C@@H](O)[C@@H]1O OVRNDRQMDRJTHS-FMDGEEDCSA-N 0.000 claims description 5
- 229950006780 n-acetylglucosamine Drugs 0.000 claims description 5
- 238000012216 screening Methods 0.000 claims description 5
- FWMNVWWHGCHHJJ-SKKKGAJSSA-N 4-amino-1-[(2r)-6-amino-2-[[(2r)-2-[[(2r)-2-[[(2r)-2-amino-3-phenylpropanoyl]amino]-3-phenylpropanoyl]amino]-4-methylpentanoyl]amino]hexanoyl]piperidine-4-carboxylic acid Chemical compound C([C@H](C(=O)N[C@H](CC(C)C)C(=O)N[C@H](CCCCN)C(=O)N1CCC(N)(CC1)C(O)=O)NC(=O)[C@H](N)CC=1C=CC=CC=1)C1=CC=CC=C1 FWMNVWWHGCHHJJ-SKKKGAJSSA-N 0.000 claims description 4
- OVRNDRQMDRJTHS-UHFFFAOYSA-N N-acelyl-D-glucosamine Natural products CC(=O)NC1C(O)OC(CO)C(O)C1O OVRNDRQMDRJTHS-UHFFFAOYSA-N 0.000 claims description 4
- MBLBDJOUHNCFQT-LXGUWJNJSA-N N-acetylglucosamine Natural products CC(=O)N[C@@H](C=O)[C@@H](O)[C@H](O)[C@H](O)CO MBLBDJOUHNCFQT-LXGUWJNJSA-N 0.000 claims description 4
- WQZGKKKJIJFFOK-RWOPYEJCSA-N beta-D-mannose Chemical compound OC[C@H]1O[C@@H](O)[C@@H](O)[C@@H](O)[C@@H]1O WQZGKKKJIJFFOK-RWOPYEJCSA-N 0.000 claims description 4
- 239000004258 Ethoxyquin Substances 0.000 claims description 3
- -1 D85 amino acids Chemical class 0.000 claims 1
- 235000001014 amino acid Nutrition 0.000 description 38
- 101000611936 Homo sapiens Programmed cell death protein 1 Proteins 0.000 description 21
- 101150107276 hpd-1 gene Proteins 0.000 description 20
- 102000048362 human PDCD1 Human genes 0.000 description 19
- 238000000034 method Methods 0.000 description 14
- 210000004027 cell Anatomy 0.000 description 11
- 238000002965 ELISA Methods 0.000 description 8
- 102220480538 Glycogen synthase kinase-3 beta_L128A_mutation Human genes 0.000 description 8
- 102220502125 Thioredoxin domain-containing protein 8_E61A_mutation Human genes 0.000 description 8
- 125000000539 amino acid group Chemical group 0.000 description 8
- 239000013078 crystal Substances 0.000 description 7
- 238000001514 detection method Methods 0.000 description 7
- 229960003301 nivolumab Drugs 0.000 description 7
- 229960002621 pembrolizumab Drugs 0.000 description 7
- 102220598780 5-hydroxytryptamine receptor 1E_K78A_mutation Human genes 0.000 description 6
- 102220502392 Putative C->U-editing enzyme APOBEC-4_E84A_mutation Human genes 0.000 description 6
- 102220484305 Thioredoxin domain-containing protein 8_D29A_mutation Human genes 0.000 description 6
- 235000018102 proteins Nutrition 0.000 description 6
- 108060003951 Immunoglobulin Proteins 0.000 description 4
- 206010028980 Neoplasm Diseases 0.000 description 4
- FAPWRFPIFSIZLT-UHFFFAOYSA-M Sodium chloride Chemical compound [Na+].[Cl-] FAPWRFPIFSIZLT-UHFFFAOYSA-M 0.000 description 4
- 238000010586 diagram Methods 0.000 description 4
- 230000033581 fucosylation Effects 0.000 description 4
- 230000013595 glycosylation Effects 0.000 description 4
- 238000006206 glycosylation reaction Methods 0.000 description 4
- 102000018358 immunoglobulin Human genes 0.000 description 4
- 230000003993 interaction Effects 0.000 description 4
- 239000000243 solution Substances 0.000 description 4
- 108010047041 Complementarity Determining Regions Proteins 0.000 description 3
- 238000013357 binding ELISA Methods 0.000 description 3
- UQLDLKMNUJERMK-UHFFFAOYSA-L di(octadecanoyloxy)lead Chemical compound [Pb+2].CCCCCCCCCCCCCCCCCC([O-])=O.CCCCCCCCCCCCCCCCCC([O-])=O UQLDLKMNUJERMK-UHFFFAOYSA-L 0.000 description 3
- 238000002474 experimental method Methods 0.000 description 3
- 108020001507 fusion proteins Proteins 0.000 description 3
- 102000037865 fusion proteins Human genes 0.000 description 3
- 238000005406 washing Methods 0.000 description 3
- WHUUTDBJXJRKMK-UHFFFAOYSA-N Glutamic acid Natural products OC(=O)C(N)CCC(O)=O WHUUTDBJXJRKMK-UHFFFAOYSA-N 0.000 description 2
- 230000015572 biosynthetic process Effects 0.000 description 2
- 201000011510 cancer Diseases 0.000 description 2
- 239000003153 chemical reaction reagent Substances 0.000 description 2
- 238000004891 communication Methods 0.000 description 2
- 238000011161 development Methods 0.000 description 2
- 238000005516 engineering process Methods 0.000 description 2
- 238000001976 enzyme digestion Methods 0.000 description 2
- 239000013604 expression vector Substances 0.000 description 2
- 230000014509 gene expression Effects 0.000 description 2
- 235000013922 glutamic acid Nutrition 0.000 description 2
- 239000004220 glutamic acid Substances 0.000 description 2
- 230000037451 immune surveillance Effects 0.000 description 2
- 238000010369 molecular cloning Methods 0.000 description 2
- 239000000178 monomer Substances 0.000 description 2
- 230000035772 mutation Effects 0.000 description 2
- 238000002360 preparation method Methods 0.000 description 2
- 102000004196 processed proteins & peptides Human genes 0.000 description 2
- 239000011780 sodium chloride Substances 0.000 description 2
- 210000004881 tumor cell Anatomy 0.000 description 2
- NFGXHKASABOEEW-UHFFFAOYSA-N 1-methylethyl 11-methoxy-3,7,11-trimethyl-2,4-dodecadienoate Chemical group COC(C)(C)CCCC(C)CC=CC(C)=CC(=O)OC(C)C NFGXHKASABOEEW-UHFFFAOYSA-N 0.000 description 1
- JKMHFZQWWAIEOD-UHFFFAOYSA-N 2-[4-(2-hydroxyethyl)piperazin-1-yl]ethanesulfonic acid Chemical compound OCC[NH+]1CCN(CCS([O-])(=O)=O)CC1 JKMHFZQWWAIEOD-UHFFFAOYSA-N 0.000 description 1
- 102100036312 5-hydroxytryptamine receptor 1E Human genes 0.000 description 1
- 108010032595 Antibody Binding Sites Proteins 0.000 description 1
- 108010074708 B7-H1 Antigen Proteins 0.000 description 1
- 102000008096 B7-H1 Antigen Human genes 0.000 description 1
- 206010006187 Breast cancer Diseases 0.000 description 1
- 208000026310 Breast neoplasm Diseases 0.000 description 1
- 241000283707 Capra Species 0.000 description 1
- 108020004705 Codon Proteins 0.000 description 1
- 108010001336 Horseradish Peroxidase Proteins 0.000 description 1
- 108010021625 Immunoglobulin Fragments Proteins 0.000 description 1
- 102000008394 Immunoglobulin Fragments Human genes 0.000 description 1
- QNAYBMKLOCPYGJ-REOHCLBHSA-N L-alanine Chemical compound C[C@H](N)C(O)=O QNAYBMKLOCPYGJ-REOHCLBHSA-N 0.000 description 1
- CKLJMWTZIZZHCS-REOHCLBHSA-N L-aspartic acid Chemical compound OC(=O)[C@@H](N)CC(O)=O CKLJMWTZIZZHCS-REOHCLBHSA-N 0.000 description 1
- ROHFNLRQFUQHCH-YFKPBYRVSA-N L-leucine Chemical compound CC(C)C[C@H](N)C(O)=O ROHFNLRQFUQHCH-YFKPBYRVSA-N 0.000 description 1
- ROHFNLRQFUQHCH-UHFFFAOYSA-N Leucine Natural products CC(C)CC(N)C(O)=O ROHFNLRQFUQHCH-UHFFFAOYSA-N 0.000 description 1
- DUKURNFHYQXCJG-UHFFFAOYSA-N Lewis A pentasaccharide Natural products OC1C(O)C(O)C(C)OC1OC1C(OC2C(C(O)C(O)C(CO)O2)O)C(NC(C)=O)C(OC2C(C(OC3C(OC(O)C(O)C3O)CO)OC(CO)C2O)O)OC1CO DUKURNFHYQXCJG-UHFFFAOYSA-N 0.000 description 1
- 206010058467 Lung neoplasm malignant Diseases 0.000 description 1
- KDXKERNSBIXSRK-UHFFFAOYSA-N Lysine Natural products NCCCCC(N)C(O)=O KDXKERNSBIXSRK-UHFFFAOYSA-N 0.000 description 1
- 239000004472 Lysine Substances 0.000 description 1
- 239000012515 MabSelect SuRe Substances 0.000 description 1
- 206010027476 Metastases Diseases 0.000 description 1
- 241000283973 Oryctolagus cuniculus Species 0.000 description 1
- 229920001030 Polyethylene Glycol 4000 Polymers 0.000 description 1
- 229920001213 Polysorbate 20 Polymers 0.000 description 1
- 108020004511 Recombinant DNA Proteins 0.000 description 1
- 239000012505 Superdex™ Substances 0.000 description 1
- 230000006044 T cell activation Effects 0.000 description 1
- 210000001744 T-lymphocyte Anatomy 0.000 description 1
- 238000002835 absorbance Methods 0.000 description 1
- 235000004279 alanine Nutrition 0.000 description 1
- 238000004458 analytical method Methods 0.000 description 1
- 230000002528 anti-freeze Effects 0.000 description 1
- 235000003704 aspartic acid Nutrition 0.000 description 1
- OQFSQFPPLPISGP-UHFFFAOYSA-N beta-carboxyaspartic acid Natural products OC(=O)C(N)C(C(O)=O)C(O)=O OQFSQFPPLPISGP-UHFFFAOYSA-N 0.000 description 1
- 210000004899 c-terminal region Anatomy 0.000 description 1
- 238000002619 cancer immunotherapy Methods 0.000 description 1
- 230000023402 cell communication Effects 0.000 description 1
- 239000006143 cell culture medium Substances 0.000 description 1
- 230000008619 cell matrix interaction Effects 0.000 description 1
- 230000005754 cellular signaling Effects 0.000 description 1
- 238000005119 centrifugation Methods 0.000 description 1
- 238000006243 chemical reaction Methods 0.000 description 1
- 239000011248 coating agent Substances 0.000 description 1
- 238000000576 coating method Methods 0.000 description 1
- 230000004154 complement system Effects 0.000 description 1
- 239000002299 complementary DNA Substances 0.000 description 1
- 125000000151 cysteine group Chemical group N[C@@H](CS)C(=O)* 0.000 description 1
- 238000007405 data analysis Methods 0.000 description 1
- 238000013461 design Methods 0.000 description 1
- MTHSVFCYNBDYFN-UHFFFAOYSA-N diethylene glycol Chemical compound OCCOCCO MTHSVFCYNBDYFN-UHFFFAOYSA-N 0.000 description 1
- 238000010790 dilution Methods 0.000 description 1
- 239000012895 dilution Substances 0.000 description 1
- 238000010494 dissociation reaction Methods 0.000 description 1
- 230000005593 dissociations Effects 0.000 description 1
- 239000012636 effector Substances 0.000 description 1
- 230000002708 enhancing effect Effects 0.000 description 1
- 239000013613 expression plasmid Substances 0.000 description 1
- 230000004927 fusion Effects 0.000 description 1
- 150000004676 glycans Chemical class 0.000 description 1
- 125000003147 glycosyl group Chemical group 0.000 description 1
- 230000036541 health Effects 0.000 description 1
- 210000004408 hybridoma Anatomy 0.000 description 1
- 230000005746 immune checkpoint blockade Effects 0.000 description 1
- 230000001900 immune effect Effects 0.000 description 1
- 230000003832 immune regulation Effects 0.000 description 1
- 210000000987 immune system Anatomy 0.000 description 1
- 230000009545 invasion Effects 0.000 description 1
- 210000004072 lung Anatomy 0.000 description 1
- 201000005202 lung cancer Diseases 0.000 description 1
- 208000020816 lung neoplasm Diseases 0.000 description 1
- 210000004962 mammalian cell Anatomy 0.000 description 1
- 230000009401 metastasis Effects 0.000 description 1
- 239000000203 mixture Substances 0.000 description 1
- 239000002808 molecular sieve Substances 0.000 description 1
- 238000005457 optimization Methods 0.000 description 1
- 230000002018 overexpression Effects 0.000 description 1
- 239000013612 plasmid Substances 0.000 description 1
- 229920001223 polyethylene glycol Polymers 0.000 description 1
- 235000010486 polyoxyethylene sorbitan monolaurate Nutrition 0.000 description 1
- 239000000256 polyoxyethylene sorbitan monolaurate Substances 0.000 description 1
- 229920001184 polypeptide Polymers 0.000 description 1
- 125000002924 primary amino group Chemical group [H]N([H])* 0.000 description 1
- 230000008569 process Effects 0.000 description 1
- 238000000746 purification Methods 0.000 description 1
- 230000009257 reactivity Effects 0.000 description 1
- 238000003259 recombinant expression Methods 0.000 description 1
- 238000011084 recovery Methods 0.000 description 1
- 238000002864 sequence alignment Methods 0.000 description 1
- 238000012163 sequencing technique Methods 0.000 description 1
- 238000002741 site-directed mutagenesis Methods 0.000 description 1
- URGAHOPLAPQHLN-UHFFFAOYSA-N sodium aluminosilicate Chemical compound [Na+].[Al+3].[O-][Si]([O-])=O.[O-][Si]([O-])=O URGAHOPLAPQHLN-UHFFFAOYSA-N 0.000 description 1
- 238000002415 sodium dodecyl sulfate polyacrylamide gel electrophoresis Methods 0.000 description 1
- 238000010561 standard procedure Methods 0.000 description 1
- 239000012089 stop solution Substances 0.000 description 1
- 238000012916 structural analysis Methods 0.000 description 1
- 230000005469 synchrotron radiation Effects 0.000 description 1
- 238000003786 synthesis reaction Methods 0.000 description 1
- 125000003396 thiol group Chemical group [H]S* 0.000 description 1
- 230000009466 transformation Effects 0.000 description 1
- 230000005747 tumor angiogenesis Effects 0.000 description 1
- 239000012224 working solution Substances 0.000 description 1
Images
Classifications
-
- C—CHEMISTRY; METALLURGY
- C07—ORGANIC CHEMISTRY
- C07K—PEPTIDES
- C07K14/00—Peptides having more than 20 amino acids; Gastrins; Somatostatins; Melanotropins; Derivatives thereof
- C07K14/435—Peptides having more than 20 amino acids; Gastrins; Somatostatins; Melanotropins; Derivatives thereof from animals; from humans
- C07K14/705—Receptors; Cell surface antigens; Cell surface determinants
-
- C—CHEMISTRY; METALLURGY
- C07—ORGANIC CHEMISTRY
- C07K—PEPTIDES
- C07K14/00—Peptides having more than 20 amino acids; Gastrins; Somatostatins; Melanotropins; Derivatives thereof
- C07K14/435—Peptides having more than 20 amino acids; Gastrins; Somatostatins; Melanotropins; Derivatives thereof from animals; from humans
- C07K14/705—Receptors; Cell surface antigens; Cell surface determinants
- C07K14/715—Receptors; Cell surface antigens; Cell surface determinants for cytokines; for lymphokines; for interferons
-
- C—CHEMISTRY; METALLURGY
- C07—ORGANIC CHEMISTRY
- C07K—PEPTIDES
- C07K16/00—Immunoglobulins [IGs], e.g. monoclonal or polyclonal antibodies
- C07K16/18—Immunoglobulins [IGs], e.g. monoclonal or polyclonal antibodies against material from animals or humans
- C07K16/28—Immunoglobulins [IGs], e.g. monoclonal or polyclonal antibodies against material from animals or humans against receptors, cell surface antigens or cell surface determinants
-
- C—CHEMISTRY; METALLURGY
- C07—ORGANIC CHEMISTRY
- C07K—PEPTIDES
- C07K16/00—Immunoglobulins [IGs], e.g. monoclonal or polyclonal antibodies
- C07K16/18—Immunoglobulins [IGs], e.g. monoclonal or polyclonal antibodies against material from animals or humans
- C07K16/28—Immunoglobulins [IGs], e.g. monoclonal or polyclonal antibodies against material from animals or humans against receptors, cell surface antigens or cell surface determinants
- C07K16/30—Immunoglobulins [IGs], e.g. monoclonal or polyclonal antibodies against material from animals or humans against receptors, cell surface antigens or cell surface determinants from tumour cells
-
- G—PHYSICS
- G01—MEASURING; TESTING
- G01N—INVESTIGATING OR ANALYSING MATERIALS BY DETERMINING THEIR CHEMICAL OR PHYSICAL PROPERTIES
- G01N33/00—Investigating or analysing materials by specific methods not covered by groups G01N1/00 - G01N31/00
- G01N33/48—Biological material, e.g. blood, urine; Haemocytometers
- G01N33/50—Chemical analysis of biological material, e.g. blood, urine; Testing involving biospecific ligand binding methods; Immunological testing
- G01N33/68—Chemical analysis of biological material, e.g. blood, urine; Testing involving biospecific ligand binding methods; Immunological testing involving proteins, peptides or amino acids
Definitions
- the present invention belongs to the field of immunology. Specifically, it relates to peptides that bind to PD-1 antibodies and uses thereof.
- Cell surface glycans play important roles in cancer, such as cell signaling and communication, tumor cell dissociation and invasion, cell-matrix interaction, tumor angiogenesis, immune regulation and metastasis formation, and immune surveillance (Varki A.( 2017). Glycobiology, 27, 3-49.). Glycosylation helps tumor cells escape immune surveillance (Okada M. et al. (2017). Cell Rep., 20, 1017-1028.) (Liu CY. et al. (2011). Proc. Natl. Acad. Sci. USA, 108, 11332-11337.) (Potapenko O.I. et al. (2010). Mol. Oncol., 4, 98-118.).
- the core part of the glycan is composed of two N-acetylglucosamine (GlcNac, NAG) and one fucose (Fucose, FUC). Fucosylation is associated with cancer (Pinho, 2015), and depleted T cells in tumors tend to be highly core-fucosylated (Okada M. et al. (2017). Cell Rep., 20, 1017-1028 ). In some cancers, such as lung and breast cancer, overexpression of core fucosylation (FUT8) has been observed (Liu CY. et al. (2011). Proc. Natl. Acad. Sci. USA, 108, 11332- 11337) (Potapenko O.I. et al.
- glycosyl groups on the surface of PD-1 may interact with anti-PD-1 antibodies, enhancing the binding activity of the two, which in turn leads to better clinical outcomes (Fessas H. et al. (2017). Semin. Oncol., 44, 136-140).
- the present invention has been completed.
- the present invention relates to the following aspects:
- a peptide preferably binding an anti-PD-1 antibody or an antigen-binding fragment thereof, said peptide comprising building block 1 and building block 2, wherein building block 1 comprises a PD-1 protein fragment selected from the group consisting of: PD
- building block 1 comprises a PD-1 protein fragment selected from the group consisting of: PD
- Structural unit 2 comprises a PD-1 protein fragment selected from the group consisting of amino acids at positions 127-133 of PD-1 protein (as shown in SEQ ID NO: 18), amino acids at positions 127-132 (show in SEQ ID NO: 19), amino acids 128-133 (shown in SEQ ID NO: 20), and amino acids 128-132 (shown in SEQ ID NO: 21).
- the peptide of claim 1, comprising structural unit 1 and structural unit 2, wherein structural unit 1 comprises amino acids at positions 29-85 of PD-1 protein (as shown in SEQ ID NO: 11) or Amino acids at positions 58-85 (as shown in SEQ ID NO: 15), and structural unit 2 comprises amino acids at positions 128-132 of PD-1 protein (as shown in SEQ ID NO: 21) or 130-132.
- Amino acids (as shown in SEQ ID NO: 3) preferably structural unit 1 comprises amino acids at positions 58 to 85 of PD-1 protein (as shown in SEQ ID NO: 15), and structural unit 2 comprises 130 of PD-1 protein - amino acid at position 132 (as shown in SEQ ID NO: 3).
- structural unit 1 comprises the sequence shown in any one of SEQ ID NO: 2 or SEQ ID NO: 10-17
- structural unit 2 comprises SEQ ID NO: 3 or The sequence shown in any one of SEQ ID NOs: 18-21, preferably structural unit 1 comprises the sequence shown in SEQ ID NO: 2, and structural unit 2 comprises the sequence shown in SEQ ID NO: 3.
- the antigen-binding fragment of the anti-PD-1 antibody is a Fab fragment, Fab', (Fab') 2 , Fab'-SH, Fab/c, Fv, Single chain antibodies (eg, scFv).
- glycosylated side chain comprises mannose, N-acetylglucosamine; fucose and ⁇ -D-mannose.
- D29, T59, E61, S62, K78, E84, D85, L128, P130, A132 or their combination of the PD-1 protein shown in SEQ ID NO: 1 is used for screening for antibodies or antigen-binding fragments thereof that bind to PD-1 use in.
- antibody refers to an immunoglobulin molecule generally composed of two pairs of polypeptide chains, each pair having one "light” (L) chain and one "heavy” (H) chain .
- Antibody light chains can be classified as kappa and lambda light chains.
- Heavy chains can be classified as mu, delta, gamma, alpha, or epsilon, and define the antibody's isotype as IgM, IgD, IgG, IgA, and IgE, respectively.
- the variable and constant regions are linked by a "J" region of about 12 or more amino acids, and the heavy chain also contains a "D" region of about 3 or more amino acids.
- Each heavy chain consists of a heavy chain variable region (VH) and a heavy chain constant region (CH).
- the heavy chain constant region consists of 3 domains (CH1, CH2 and CH3).
- Each light chain consists of a light chain variable region (VL) and a light chain constant region (CL).
- the light chain constant region consists of one domain, CL.
- the constant regions of the antibodies mediate the binding of the immunoglobulin to host tissues or factors, including various cells of the immune system (eg, effector cells) and the first component (Clq) of the classical complement system.
- the VH and VL regions can also be subdivided into regions of high variability called complementarity determining regions (CDRs) interspersed with more conserved regions called framework regions (FRs).
- CDRs complementarity determining regions
- Each VH and VL consists of 3 CDRs and 4 FRs arranged in the following order: FR1, CDR1, FR2, CDR2, FR3, CDR3, FR4 from amino terminus to carboxy terminus.
- the assignment of amino acids to regions or domains follows the Kabat Sequences of Proteins of Immunological Interest (National Institutes of Health, Bethesda, Md. (1987 and 1991)), or Chothia & Lesk (1987) J. Mol. Biol. 196:901 -917; definition by Chothia et al. (1989) Nature 342:878-883.
- antibody is not limited by any particular method of producing an antibody.
- it includes, in particular, recombinant antibodies, monoclonal antibodies and polyclonal antibodies.
- Antibodies can be of different isotypes, eg, IgG (eg, IgGl, IgG2, IgG3, or IgG4 subtype), IgAl, IgA2, IgD, IgE, or IgM antibodies.
- the terms “monoclonal antibody” and “monoclonal antibody” refer to an antibody or a fragment of an antibody from a population of highly homologous antibody molecules, that is, excluding natural mutations that may arise spontaneously, A population of identical antibody molecules.
- Monoclonal antibodies are highly specific for a single epitope on an antigen.
- Polyclonal antibodies are relative to monoclonal antibodies, which generally comprise at least two or more different antibodies that generally recognize different epitopes on an antigen.
- Monoclonal antibodies are typically obtained using the hybridoma technology first reported by Kohler et al. (Nature, 256:495, 1975), but can also be obtained using recombinant DNA technology (eg, see U.S. Patent 4,816,567).
- humanized antibody refers to the replacement of all or part of the CDR regions of a human immunoglobulin (acceptor antibody) with the CDR regions of a non-human antibody (donor antibody)
- the antibody or antibody fragment of which the donor antibody can be a non-human (eg, mouse, rat or rabbit) antibody with the desired specificity, affinity or reactivity.
- some amino acid residues in the framework region (FR) of the acceptor antibody can also be replaced by amino acid residues of corresponding non-human antibodies, or by amino acid residues of other antibodies, to further improve or optimize the performance of the antibody.
- antigen-binding fragments include, but are not limited to: Fab fragments, Fab', (Fab') 2 , Fab/c, Fv, Fab'-SH, single chain antibodies (eg, scFv).
- Fab fragment consists of a light chain and the variable regions of CH1 and a heavy chain.
- the heavy chain of a Fab molecule cannot form a disulfide bond with another heavy chain molecule.
- Fab'fragment contains part of one light chain and one heavy chain (which contains the VH and CH1 domains and also the region between the CH1 and CH2 domains) part) so that an interchain disulfide bond can be formed between the two heavy chains of the two Fab' fragments to form an F(ab') 2 molecule.
- F(ab') 2 fragment contains two light chains and two heavy chains containing part of the constant region between the CH1 and CH2 domains, so that the Interchain disulfide bonds are formed between the chains.
- the F(ab') 2 fragment thus consists of two Fab' fragments held together by disulfide bonds between the two heavy chains.
- Fv region includes variable regions from heavy and light chains, but lacks constant regions.
- Fab'-SH is the designation herein for Fab' wherein one or more cysteine residues of the constant domains carry free thiol groups.
- Figure 1 Overall structural model diagram of PD-1 protein complex with 14C12H1L1-Fab.
- FAB-HC 14C12H1L1-Fab heavy chain portion;
- FAB-LC 14C12H1L1-Fab light chain portion.
- Figure 2 Schematic diagram of the interaction between PD-1 protein and 14C12H1L1-Fab heavy chain amino acids.
- FAB-HC 14C12H1L1-Fab heavy chain portion.
- Figure 3 Schematic diagram of the interaction between PD-1 protein and 14C12H1L1-Fab light chain amino acids.
- FAB-LC 14C12H1L1-Fab light chain portion.
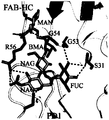
- Figure 4 Schematic diagram of the interaction between the glycosylated side chain of PD-1 protein Asn58 and the heavy chain amino acids of 14C12H1L1-Fab.
- MAN mannose, mannose
- NAG N Acetyl Glucosamine, acetylglucosamine
- FUC Fucose, fucose
- BMA ⁇ -D-mannose, ⁇ -D-mannose.
- FAB-HC 14C12H1L1-Fab heavy chain portion
- FAB-LC 14C12H1L1-Fab light chain portion.
- Figure 5 Binding surface of PD-1 to 14C12H1L1-Fab, Pembrolizumab or nivolumab.
- the residues in contact with 14C12H1L1-Fab are E61, S62 amino acid residues
- the residues in contact with nivolumab are V64, N66, Y68, Q75, T76, D77, K78, P83 amino acid residues
- the overlapping residue bound by 14C12H1L1-Fab and nivolumab is the amino acid residue at position E84.
- the residues in contact with pembrolizumab are L128, A129 amino acid residues, and the overlapping residues bound by 14C12H1L1-FAB and pembrolizumab are T59, P130, A131, K132 amino acid residues.
- the amino acid residue at position D85 was not shown due to the occlusion of the steric structure.
- Figure 6 The binding activity of anti-PD-1 antibody to human PD-1 protein mutant hPD1(D29A)-mFc and hPD1(E61A)-mFc was detected by combined ELISA method.
- Figure 7 The binding activity of anti-PD-1 antibody to human PD-1 protein mutant hPD1(K78A)-mFc and hPD1(E84A)-mFc was detected by combined ELISA method.
- Figure 8 Binding ELISA method to detect the binding activity of anti-PD-1 antibody to human PD-1 protein mutant hPD1(L128A)-mFc.
- Figure 9 Detection results of kinetic parameters of the binding of anti-PD-1 antibody to human PD-1 protein hPD1-mFc.
- Fig. 10 The detection results of kinetic parameters of the binding of anti-PD-1 antibody to human PD-1 protein mutant hPD1(D29A)-mFc.
- Figure 11 The results of the kinetic parameters of the binding of anti-PD-1 antibody to human PD-1 protein mutant hPD1(E61A)-mFc.
- Figure 12 The results of the kinetic characteristic parameters of the binding of anti-PD-1 antibody to human PD-1 protein mutant hPD1(K78A)-mFc.
- Figure 13 The results of the kinetic parameter detection of anti-PD-1 antibody binding to human PD-1 protein mutant hPD1(E84A)-mFc.
- Fig. 14 The detection results of kinetic parameters of the binding of anti-PD-1 antibody to human PD-1 protein mutant hPD1(L128A)-mFc.
- hPD1-mFc produced by Kangfang Bio, batch number: 20181025 (Genbank ID of hPD-1: NP_005009, Genbank ID of mFc: P01863).
- hPD1(D29A)-mFc produced by Kangfang Biotechnology, batch number 20191113.
- hPD1(E61A)-mFc produced by Kangfang Biotechnology, batch number 20191113.
- hPD1(K78A)-mFc produced by Kangfang Biotechnology, batch number 20191113.
- hPD1(E84A)-mFc produced by Kangfang Biotechnology, batch number 20191113.
- hPD1(L128A)-mFc produced by Kangfang Biotechnology, batch number 20191113.
- the sequence of human PD-1 was searched through the NCBI protein database (PD-1 Genbank ID: NP_005009), and the amino acid sequence of the extracellular region of human PD-1 (amino acids 1-170) was combined with his purification tag sequence (SEQ ID NO: 6 ) for fusion design, and the fusion protein was abbreviated as "PD1-his” (SEQ ID NO: 7), also expressed as "hPD1-his”.
- the cDNA sequence encoding the fusion protein was derived from the amino acid codon optimization and gene synthesis commissioned by Nanjing GenScript Biotechnology. Referring to the standard techniques introduced in "Molecular Cloning Experiment Guide (Second Edition)", PCR, enzyme digestion, gel recovery, Standard molecular cloning techniques such as ligation transformation, colony PCR or enzyme digestion identification are used to subclon the target gene into a mammalian cell expression vector, and the target gene of the recombinant expression vector is further sequenced and analyzed. After the sequencing was verified to be correct, the endotoxin-free expression plasmid was prepared in large quantities and the plasmid was transiently transfected into HEK293 cells for protein expression. After 7 days of culture, the cell culture medium was collected and affinity purified using MabSelect SuRe column (GE Healthcare).
- the inventors based on the reported PD-1-nivolumab Fab (Tan SH, et al. (2017). Nat Commun., 8, 14369) and PD-1-Pembrolizumab Fab (Horita S et al. (2016). Sci Rep. ., 6, 35297) complex structure, using the method of X-ray crystal diffraction, to The Fab portion of antibody 14C12H1L1 (14C12H1L1-Fab) interacted with the PD-1 antigen at the resolution of 14C12H1L1 and compared the differences among the three.
- amino acid sequence of the heavy chain portion of the antibody 14C12H1L1-Fab is as follows, the underlined portion is CH1, the bold bold is the CDR region, and the underlined bold italic is his-tag:
- amino acid sequence of the light chain in the antibody 14C12H1L1-Fab is as follows, the underlined part is CL, and the bolded bold is the CDR region:
- 14C12H1L1-Fab binds to PD-1 in a manner distinct from pembrolizumab and nivolumab. Although all three block the binding between PD-1 and PD-L1, according to literature reports, pembrolizumab and nivolumab bind to PD-1 in a glycosylation-independent manner (Tan SH, et al. (2017). ).Nat Commun., 8, 14369.) and PD-1-Pembrolizumab Fab (Horita S et al. (2016). Sci Rep., 6, 35297), 14C12H1L1-Fab showed on the BC loop of PD-1 Numerous interactions with sugar side chains linked at the N58 position.
- PD1 protein such as PD-1-His
- 14C12H1L1-Fab-his was mixed with 14C12H1L1-Fab-his at a molar ratio of 1:2, and incubated on ice for 2 hours.
- the mixture was then purified by molecular sieves (Superdex 200 10/300 column, GE Healthcare) to obtain a complex of PD-1 protein and 14C12H1L1-Fab-his using buffer conditions of 20 mM HEPEs, pH 7.5, 100 mM NaCl, 5 mM DTT.
- the complex peak was collected and concentrated by centrifugation (Millipore, MWCO 10 kDa), and the final complex concentration was about 10 mg/ml.
- the concentrated complex was used for the primary screening of crystals and kept in a crystal room at 20°. Two weeks later, the crystal plate was observed under a microscope, and the crystallographic conditions were selected to repeat and optimize the crystallographic conditions.
- the crystals were grown in PEG II suit F2 (0.1M MES, pH 6.5, 20% PEG4000, 0.6M sodium chloride) with 30% EG antifreeze. SDS-PAGE showed that the crystal contents were PD1-His+14C12H1L1-Fab-His complex. After acquiring available crystals, the resolution data was collected by using the Shanghai Synchrotron Radiation Light Source for diffraction to diffraction pattern.
- the data analysis process is as follows: DIALS is used to index and integrate the data. Aimless was used to analyze and merge the processed data, and 5% of the data were randomly selected for Rfree estimation.
- the Molrep molecular replacement procedure was used to find the phase solution in two steps. After sequence alignment, the sequence homology between 14C12H1L1-Fab and 6foe in the PDB database is as high as 85%, so use 6foe to find the solution of 14C12H1L1-Fab-his, then fix the position of 14C12H1L1-Fab-his, and use PD-1 monomer The structure of (PDB: 3rre) to find the solution of PD-1.
- the analytical result is that an asymmetric unit contains a PD1-his monomer and a 14C12H1L1-Fab-his fragment.
- the model is then revised in the reciprocal space using REFMAC5.
- the protein model was corrected in real space using COOT.
- the model is in good agreement with the electron density map, the crystallographic R-factor and Rfree are 0.21 and 0.27, respectively (Fig. 5), and the stereochemical parameters of the structural model are within a reasonable range.
- 5 amino acid site mutations of human PD-1 protein were selected as 29 aspartic acid, 61 glutamic acid, 78 lysine, 84 glutamic acid and 128 leucine. is alanine [Lee, J.Y., et al., Structural basis of checkpoint blockade by monoclonal antibodies in cancer immunotherapy.
- Dilute PD-1 antigen to 1 ⁇ g/mL with coating buffer add 50 ⁇ L per well to the ELISA plate, and incubate at 4°C overnight. After washing the plate with PBST, 300 ⁇ L of 1% BSA (PBS) was added to each well, and the plate was blocked at 37° C. for 2 h.
- PBS 1% BSA
- the anti-PD-1 antibody 14C12H1L1(hG1TM) was diluted with PBST to 0.3333 ⁇ g/mL as the starting concentration, and diluted 1:3 down to 0.1111 ⁇ g/mL, 0.0370 ⁇ g/mL on the ELISA plate , 0.0123 ⁇ g/mL, 0.0041 ⁇ g/mL, 0.0014 ⁇ g/mL, 0.0005 ⁇ g/mL, a total of 7 gradient concentrations, a blank control was set up, and 2 duplicate wells were made, each well 100 ⁇ L, mixed and incubated at 37°C 30min.
- the detection results are shown in Table 1 and Figures 6-8.
- the five human PD-1 protein mutants D29A, E61A, K78A, E84A, L128A bind to the anti-PD-1 antibody with EC50 of 0.021nM, 2.47 ⁇ 10 12 nM, 0.023nM, 0.022nM, 4.207nM.
- the binding ability of E61A and L128A mutants to anti-PD-1 antibody was significantly reduced.
- the anti-PD-1 antibody was diluted to 5 ⁇ g/mL with PBS (containing 0.02% Tween-20, 0.1% BSA, pH 7.4) and then immobilized on the surface of AHC sensor (Fortebio) for 120 s, and the sensor was equilibrated in the buffer
- the anti-PD-1 antibody immobilized on the sensor binds to each PD1-mFc mutant at a concentration of 1.24-100nM (three-fold dilution) for 120s, and the protein dissociates in the buffer for 300s.
- the detection temperature was 37° C.
- the detection frequency was 0.3 Hz
- the vibration rate of the sample plate was 1000 rpm. Data were analyzed with a 1:1 model fit to yield affinity constants.
- the binding results of anti-PD-1 antibody and each PD-1 mutant are shown in Figure 9-14 and Table 2.
- Five human PD-1 protein mutants D29A, E61A, K78A, E84A, L128A bind to anti-PD-1 antibody
- the affinity constants were 4.25E-10M, 3.30E-08M, 1.31E-10M, 6.65E-10M, 7.68E-09M, respectively. Consistent with the binding ELISA results, the binding ability of the two mutants E61A and L128A to anti-PD-1 antibodies was significantly reduced.
Landscapes
- Health & Medical Sciences (AREA)
- Life Sciences & Earth Sciences (AREA)
- Chemical & Material Sciences (AREA)
- Organic Chemistry (AREA)
- Immunology (AREA)
- Molecular Biology (AREA)
- Proteomics, Peptides & Aminoacids (AREA)
- General Health & Medical Sciences (AREA)
- Biochemistry (AREA)
- Medicinal Chemistry (AREA)
- Biophysics (AREA)
- Cell Biology (AREA)
- Genetics & Genomics (AREA)
- Engineering & Computer Science (AREA)
- Biomedical Technology (AREA)
- Gastroenterology & Hepatology (AREA)
- Zoology (AREA)
- Hematology (AREA)
- Urology & Nephrology (AREA)
- Toxicology (AREA)
- Physics & Mathematics (AREA)
- Pathology (AREA)
- General Physics & Mathematics (AREA)
- Biotechnology (AREA)
- Analytical Chemistry (AREA)
- Food Science & Technology (AREA)
- Microbiology (AREA)
- Peptides Or Proteins (AREA)
Abstract
提供了结合抗PD-1抗体或其抗原结合片段的肽,其包含结构单元1和2,其中结构单元1包含PD-1蛋白的58位-85位的氨基酸,结构单元2包含PD-1蛋白的130-132位的氨基酸。
Description
本发明属于免疫学领域。具体涉及结合PD-1抗体的肽及其应用。
细胞表面聚糖在癌症中起着重要作用,例如细胞信号和通讯,肿瘤细胞的解离和侵袭,细胞与基质的相互作用,肿瘤血管生成,免疫调节和转移形成以及免疫监测(Varki A.(2017).Glycobiology,27,3-49.)。糖基化有助于肿瘤细胞逃脱免疫监视(Okada M.et al.(2017).Cell Rep.,20,1017-1028.)(Liu CY.et al.(2011).Proc.Natl.Acad.Sci.USA,108,11332-11337.)(Potapenko O.I.et al.(2010).Mol.Oncol.,4,98-118.)。
PD-1的细胞外免疫球蛋白可变(IgV)域中有四个报道的糖基化位点,分别是N49,N58,N74和N116(Chen D.et al.(2019).iScience,14,113-124.)(Na Z.et al.(2017).Cell Res.,27,147-150.)(Okada M.et al.(2017).Cell Rep.,20,1017-1028)。据报道,在哺乳动物(Tan SH.et al.(2017).Nat Commun.,8,14369)细胞的PD-1的N58氨基酸残基位于PD-1的BC环上,其糖基化程度很高,多数情况下其核心部分的聚糖是由两个N-乙酰氨基葡萄糖(GlcNac,NAG)和一个岩藻糖(Fucose,FUC)组成。岩藻糖糖基化与癌症有关(Pinho,2015),肿瘤中耗竭T细胞往往为高度核心岩藻糖基化的(Okada M.et al.(2017).Cell Rep.,20,1017-1028)。在一些癌症,例如肺癌和乳腺癌中,观察到核心岩藻糖基化(FUT8)的过表达(Liu CY.et al.(2011).Proc.Natl.Acad.Sci.USA,108,11332-11337)(Potapenko O.I.et al.(2010).Mol.Oncol.,4,98-118)。核心岩藻糖基化的丧失可降低细胞表面PD-1的表达,进而增强T细胞活化(Okada M.et al.(2017).Cell Rep.,20,1017-1028)。
PD-1表面的糖基可能会与抗PD-1抗体相互作用,增强的两者的结合活性,进而表现为更好的临床结果(Fessas H.et al.(2017).Semin.Oncol.,44,136-140)。
发明内容
本发明人发现,PD-1蛋白(PD-1Genbank ID:NP_005009,SEQ ID NO:1)的T59、E61、S62、E84、D85位氨基酸与抗PD-1抗体例如14C12H1L1-Fab重链的Y32、D33、S52、G54、Y57、Y100位氨基酸,PD-1蛋白的P130、K131、A132位氨基酸与抗PD-1抗体例如14C12H1L1-Fab轻链的F325、E324、D323位氨基酸发生相互作用。由此完成了本发明。
具体地,本发明涉及以下几个方面:
1.肽,优选结合抗PD-1抗体或其抗原结合片段,所述肽包含结构单元1和结构单元2,其中结构单元1包含选自以下各项组成的组的PD-1蛋白片段:PD-1蛋白的28位-86位的氨基酸(如SEQ ID NO:10所示),28位-85位的氨基酸(如SEQ ID NO:11所示),29位-86位的氨基酸(如SEQ ID NO:12所示),29位-85位的氨基酸(如SEQ ID NO:13所示),58位-86位的氨基酸(如SEQ ID NO:14所示)、58位-85位的氨基酸(如SEQ ID NO:15所示),59位-86位的氨基酸(如SEQ ID NO:16所示)和59位-85位的氨基酸(如SEQ ID NO:17所示),
结构单元2包含选自以下各项组成的组的PD-1蛋白片段:PD-1蛋白的127位-133位的氨基酸(如SEQ ID NO:18所示),127位-132位的氨基酸(如SEQ ID NO:19所示),128位-133位的氨基酸(如SEQ ID NO:20所示),和128位-132位的氨基酸(如SEQ ID NO:21所示)。
2.权利要求1所述的肽,所述肽包含结构单元1和结构单元2,其中结构单元1包含PD-1蛋白的29位-85位的氨基酸(如SEQ ID NO:11所示)或58位-85位的氨基酸(如SEQ ID NO:15所示),结构单元2包含PD-1蛋白的128位-132位的氨基酸(如SEQ ID NO:21所示)或130-132位的氨基酸(如SEQ ID NO:3所示),优选结构单元1包含PD-1蛋白的58位-85位的氨基酸(如SEQ ID NO:15所示),结构单元2包含PD-1蛋白的130-132位的氨基酸(如SEQ ID NO:3所示)。
3.项目1或2所述的肽,其中PD-1蛋白的氨基酸序列如SEQ ID NO:1所示。
4.项目1-3任一项所述的肽,其中结构单元1包含SEQ ID NO:2 或SEQ ID NO:10-17任一项所示的序列,结构单元2包含SEQ ID NO:3或SEQ ID NO:18-21任一项所示的序列,优选结构单元1包含SEQ ID NO:2所示的序列,结构单元2包含SEQ ID NO:3所示的序列。
5.项目1-4任一项所述的肽,其中所述抗PD-1抗体的抗原结合片段为Fab片段、Fab’、(Fab’)
2、Fab’-SH、Fab/c、Fv、单链抗体(例如,scFv)。
6.项目5所述的肽,其中所述Fab片段的重链氨基酸序列如SEQ ID NO:4所示,轻链氨基酸序列如SEQ ID NO:5所示。
7.项目1-6任一项所述的肽,其中PD-1蛋白的T59、E61、S62、E84、D85氨基酸与抗PD-1抗体(例如14C12H1L1-Fab)重链的Y32、D33、S52、G54、Y57、Y100位氨基酸结合,PD-1蛋白的P130、K131、A132氨基酸与抗PD-1抗体轻链的F325、E324、D323位氨基酸结合,PD-1蛋白的N58糖基化侧链与抗PD-1抗体(例如14C12H1L1-Fab)重链氨基酸(例如S31、G53、G54和R56)结合。
8.项目7所述的肽,其中所述糖基化侧链包含甘露糖,N-乙酰氨基葡萄糖;岩藻糖和β-D-甘露糖。
9.项目1-8任一项所述的肽在筛选结合PD-1的抗体或其抗原结合片段中的用途。
10.SEQ ID NO:1所示的PD-1蛋白的D29,T59,E61,S62,K78,E84,D85,L128,P130,A132或其组合在筛选结合PD-1的抗体或其抗原结合片段中的用途。
如本文中所使用的,术语“抗体”是指,是指通常由两对多肽链(每对具有一条“轻”(L)链和一条“重”(H)链)组成的免疫球蛋白分子。抗体轻链可分类为κ和λ轻链。重链可分类为μ、δ、γ、α或ε,并且分别将抗体的同种型定义为IgM、IgD、IgG、IgA和IgE。在轻链和重链内,可变区和恒定区通过大约12或更多个氨基酸的“J”区连接,重链还包含大约3个或更多个氨基酸的“D”区。各重链由重链可变区(VH)和重链恒定区(CH)组成。重链恒定区由3个结构域(CH1、CH2和CH3)组成。各轻链由轻链可变区(VL)和轻链恒定区(CL)组成。轻链恒定区由一个结构 域CL组成。抗体的恒定区可介导免疫球蛋白与宿主组织或因子,包括免疫系统的各种细胞(例如,效应细胞)和经典补体系统的第一组分(C1q)的结合。VH和VL区还可被细分为具有高变性的区域(称为互补决定区(CDR)),其间散布有较保守的称为构架区(FR)的区域。各VH和VL由按下列顺序:FR1、CDR1、FR2、CDR2、FR3、CDR3、FR4从氨基末端至羧基末端排列的3个CDR和4个FR组成。各重链/轻链对的可变区(VH和VL)分别形成抗体结合部位。氨基酸至各区域或结构域的分配遵循Kabat Sequences of Proteins of Immunological Interest(National Institutes of Health,Bethesda,Md.(1987 and 1991)),或Chothia & Lesk(1987)J.Mol.Biol.196:901-917;Chothia等人(1989)Nature 342:878-883的定义。术语“抗体”不受任何特定的产生抗体的方法限制。例如,其包括,特别地,重组抗体、单克隆抗体和多克隆抗体。抗体可以是不同同种型的抗体,例如,IgG(例如,IgG1,IgG2,IgG3或IgG4亚型),IgA1,IgA2,IgD,IgE或IgM抗体。
如本文中所使用的,术语“单抗”和“单克隆抗体”是指,来自一群高度同源的抗体分子中的一个抗体或抗体的一个片段,也即除可能自发出现的自然突变外,一群完全相同的抗体分子。单抗对抗原上的单一表位具有高特异性。多克隆抗体是相对于单克隆抗体而言的,其通常包含至少2种或更多种的不同抗体,这些不同的抗体通常识别抗原上的不同表位。单克隆抗体通常可采用Kohler等首次报道的杂交瘤技术获得(Nature,256:495,1975),但也可采用重组DNA技术获得(如参见U.S.Patent 4,816,567)。
如本文中所使用的,术语“人源化抗体”是指,人源免疫球蛋白(受体抗体)的全部或部分CDR区被一非人源抗体(供体抗体)的CDR区替换后得到的抗体或抗体片段,其中的供体抗体可以是具有预期特异性、亲和性或反应性的非人源(例如,小鼠、大鼠或兔)抗体。此外,受体抗体的构架区(FR)的一些氨基酸残基也可被相应的非人源抗体的氨基酸残基替换,或被其他抗体的氨基酸残基替换,以进一步完善或优化抗体的性能。关于人源化抗体的更多详细内容,可参见例如,Jones et al.,Nature,321:522525(1986);Reichmann et al.,Nature,332:323 329(1988);Presta,Curr.Op. Struct.Biol.,2:593 596(1992);和Clark,Immunol.Today 21:397 402(2000)。
在本发明中,抗原结合片段包括,但不限于:Fab片段、Fab’、(Fab’)
2、Fab/c、Fv、Fab’-SH、单链抗体(例如,scFv)。
如本发明中所使用的,术语“Fab片段”由一条轻链和C
H1以及一条重链的可变区组成。Fab分子的重链不能与另一条重链分子形成二硫键。
如本发明中所使用的,术语“Fab’片段”含有一条轻链和一条重链的部分(其含有V
H结构域和C
H1结构域以及还有C
H1与C
H2结构域之间的区域的部分),以便可在两个Fab’片段的两条重链之间形成链间二硫键以形成F(ab’)
2分子。
如本发明中所使用的,术语“F(ab’)
2片段”含有两条轻链和两条含有C
H1与C
H2结构域之间的恒定区的部分的重链,以便在两条重链之间形成链间二硫键。F(ab’)
2片段从而由通过两条重链之间的二硫键保持在一起的两个Fab’片段组成。
如本发明中所使用的,术语“Fv区”包含来自重链和轻链的可变区,但缺乏恒定区。
如本发明中所使用的,术语“Fab’-SH”是本文对Fab’的命名,其中恒定结构域的一个或多个半胱氨酸残基携带游离硫醇基团。
图1:PD-1蛋白与14C12H1L1-Fab复合物的整体结构模型图。FAB-HC:14C12H1L1-Fab重链部分;FAB-LC:14C12H1L1-Fab轻链部分。
图2:PD-1蛋白与14C12H1L1-Fab重链氨基酸相互作用示意图。FAB-HC:14C12H1L1-Fab重链部分。
图3:PD-1蛋白与14C12H1L1-Fab轻链氨基酸相互作用示意图。FAB-LC:14C12H1L1-Fab轻链部分。
图4:PD-1蛋白Asn58糖基化侧链与14C12H1L1-Fab重链氨基酸相互作用示意图。MAN:mannose,甘露糖;NAG:N Acetyl Glucosamine,乙酰氨基葡萄糖;FUC:Fucose,岩藻糖;BMA:β-D-mannose,β-D-甘露糖。FAB-HC:14C12H1L1-Fab重链部分;FAB-LC:14C12H1L1-Fab轻链部分。
图5:PD-1与14C12H1L1-Fab,Pembrolizumab或nivolumab的结合表面。PD-1蛋白中,与14C12H1L1-Fab接触的残基是E61、S62位氨基酸残基,而与nivolumab接触的残基是V64、N66、Y68、Q75、T76、D77、K78、P83位氨基酸残基,由14C12H1L1-Fab和nivolumab结合的重叠残基是E84位氨基酸残基。与pembrolizumab接触的残基是L128、A129位氨基酸残基,由14C12H1L1-FAB和pembrolizumab结合的重叠残基是T59、P130、A131、K132位氨基酸残基。D85位氨基酸残基由于立体结构遮挡未能显示。
图6:结合ELISA方法检测抗PD-1抗体与人PD-1蛋白突变体hPD1(D29A)-mFc、hPD1(E61A)-mFc的结合活性。
图7:结合ELISA方法检测抗PD-1抗体与人PD-1蛋白突变体hPD1(K78A)-mFc、hPD1(E84A)-mFc的结合活性。
图8:结合ELISA方法检测抗PD-1抗体与人PD-1蛋白突变体hPD1(L128A)-mFc的结合活性。
图9:抗PD-1抗体与人PD-1蛋白hPD1-mFc结合的动力学特征参数检测结果。
图10:抗PD-1抗体与人PD-1蛋白突变体hPD1(D29A)-mFc结合的动力学特征参数检测结果。
图11:抗PD-1抗体与人PD-1蛋白突变体hPD1(E61A)-mFc结合的动力学特征参数检测结果。
图12:抗PD-1抗体与人PD-1蛋白突变体hPD1(K78A)-mFc结合的动力学特征参数检测结果。
图13:抗PD-1抗体与人PD-1蛋白突变体hPD1(E84A)-mFc结合的动力学特征参数检测结果。
图14:抗PD-1抗体与人PD-1蛋白突变体hPD1(L128A)-mFc结合的动力学特征参数检测结果。
下面将结合实施例对本发明的实施方案进行详细描述。本领域技术人员将会理解,下面的实施例仅用于说明本发明,而不应视为限定本发明的 范围。实施例中未注明具体技术或条件者,按照本领域内的文献所描述的技术或条件或按照产品说明书进行。所用试剂或仪器未注明生产厂商者,为可以通过市场购买获得的常规产品。
主要仪器与试剂
PD-1-his(康方生物生产)
14C12H1L1-Fab-his(康方生物生产)
14C12H1L1(hG1TM)(康方生物生产)
hPD1-mFc:康方生物生产,批号:20181025(hPD-1的Genbank ID:NP_005009,mFc的Genbank ID:P01863)。
hPD1(D29A)-mFc:康方生物生产,批号20191113。
hPD1(E61A)-mFc:康方生物生产,批号20191113。
hPD1(K78A)-mFc:康方生物生产,批号20191113。
hPD1(E84A)-mFc:康方生物生产,批号20191113。
hPD1(L128A)-mFc:康方生物生产,批号20191113。
制备例1.人PD1-his融合蛋白的制备
通过NCBI蛋白质数据库查找人PD-1的序列(PD-1Genbank ID:NP_005009),将人PD-1胞外区氨基酸序列(1位氨基酸-170位氨基酸)与his纯化标签序列(SEQ ID NO:6)进行融合设计,融合蛋白简写命名为“PD1-his”(SEQ ID NO:7),也表示为“hPD1-his”。
编码融合蛋白的cDNA序列来源于委托南京金斯瑞生物进行的氨基酸密码子优化和基因合成,参照《分子克隆实验指南(第二版)》介绍的标准技术,采用PCR、酶切、胶回收、连接转化、菌落PCR或酶切鉴定等标准的分子克隆技术将目的基因亚克隆到哺乳动物细胞表达载体,并进一步对重组表达载体的目的基因进行测序分析。测序验证正确后,中大量制备去内毒素级别的表达质粒并将质粒瞬时转染HEK293细胞进行蛋白表达。培养7天后收集细胞培养液,采用MabSelect SuRe柱料(GE Healthcare)进行亲和纯化。
实施例1.人PD-1抗原与抗PD-1抗体结合表位分析
发明人基于已报道的PD-1-nivolumab Fab(Tan SH,et al.(2017).Nat Commun.,8,14369)和PD-1-Pembrolizumab Fab(Horita S et al.(2016).Sci Rep.,6,35297)的复杂结构,采用X线晶体衍射的方法,以
的分辨率研究了抗体14C12H1L1的Fab部分(14C12H1L1-Fab)和PD-1抗原相互作用,并比较了三者的差异。
抗体14C12H1L1-Fab中的重链部分的氨基酸序列如下,下划线部分为CH1,加粗黑体的为CDR区域,下划线粗斜体的为组氨酸标签(his-tag):
抗体14C12H1L1-Fab中的轻链的氨基酸序列如下,下划线部分为CL,加粗黑体的为CDR区域:
发明人出乎意料地发现,14C12H1L1-Fab通过区别于pembrolizumab和nivolumab的方式与PD-1结合。尽管三者都阻断了PD-1和PD-L1之间的结合,但是,根据文献报道,pembrolizumab和nivolumab以不依赖糖基化的方式结合至PD-1(Tan SH,et al.(2017).Nat Commun.,8,14369.)和PD-1-Pembrolizumab Fab(Horita S et al.(2016).Sci Rep.,6,35297),14C12H1L1-Fab在PD-1的BC环上显示出与N58位链接的糖侧链的大量相互作用。
实验方法:将PD1蛋白,如PD-1-His,与14C12H1L1-Fab-his按照1∶2摩尔比混合,冰上孵育2小时。混合物随后进行分子筛(Superdex 200 10/300 column,GE Healthcare)纯化得到PD-1蛋白与14C12H1L1-Fab-his的复合物,所使用的缓冲液条件为20mM HEPEs,pH 7.5,100mM NaCl,5mM DTT。收取复合物峰进行离心浓缩(Millipore,MWCO 10kDa),最终复合物浓度约为10mg/ml。将浓缩后的复合物用于晶体初筛,于20°晶体房静置。两周后,在显微镜下观察晶体板,选取晶体外形较好的条件进行结晶条件的重复及优化。晶体的生长条件为PEG II suit F2(0.1M MES,pH 6.5,20%PEG4000,0.6M sodium chloride),防冻剂为30%EG。SDS-PAGE显示晶体内容物为PD1-His+14C12H1L1-Fab-His复合物。在获取可用晶体后,通过使用上海同步辐射光源收集分辨率数据为衍射到
的衍射图形。
数据解析流程如下:采用DIALS对数据进行指标化、积分处理。用Aimless分析、合并处理数据,随机选取5%数据进行Rfree估测。采用Molrep分子置换程序分两步来寻找相位解。经过序列比对,14C12H1L1-Fab与PDB数据库中6foe的序列同源性高达85%,所以用6foe寻找14C12H1L1-Fab-his的解,然后固定14C12H1L1-Fab-his的位置,用PD-1单体的结构(PDB:3rre)来寻找PD-1的解。解析结果为一个不对称单位里包含一个PD1-his单体和一个14C12H1L1-Fab-his片段。随后采用REFMAC5在倒易空间中进行模型修正。采用COOT对蛋白模型在实空间进行修正。模型与电子密度图吻合良好,晶体学R因子和Rfree分别为0.21和0.27(图5),结构模型的立体化学参数处在合理范围内。
结构分析结果如图1-5所示。
实施例2.抗PD-1抗体14C12H1L1(hG1TM)抗原结合表位的氨基酸定点突变研究
根据Nivolumab抗体结合位点,选择了人PD-1蛋白29位天冬氨酸、61位谷氨酸、78位赖氨酸、84位谷氨酸、128位亮氨酸5个氨基酸位点突变为丙氨酸[Lee,J.Y.,et al.,Structural basis of checkpoint blockade by monoclonal antibodies in cancer immunotherapy.Nature Communications,2016.7(1):p.13354.],这些突变体分别记为hPD1(D29A)-mFc、hPD1(E61A)-mFc、hPD1(K78A)-mFc、hPD1(E84A)-mFc、hPD1(L128A)-mFc采用结合ELISA以及Fortebio Kinetics方法检测抗PD-1抗体与这些突变 体的结合活性,实验用未突变的PD1-mFc蛋白作为对照对比。
1、结合ELISA方法检测抗PD-1抗体14C12H1L1(hG1TM)与人PD-1蛋白突变体的结合活性
用包被缓冲液将PD-1抗原稀释成1μg/mL,每孔50μL加入酶标板中,置于4℃孵育过夜。用PBST洗板后每孔加入300μL 1%BSA(PBS),37℃封闭2h。洗板后用PBST将抗PD-1抗体(14C12H1L1(hG1TM))稀释成0.3333μg/mL作为起始浓度,在酶标板上进行1∶3往下稀释为0.1111μg/mL,0.0370μg/mL,0.0123μg/mL,0.0041μg/mL,0.0014μg/mL,0.0005μg/mL共7个梯度浓度,另设空白对照,均做2个复孔,每孔100μL,混匀后置于37℃孵育30min。洗板后每孔加入50μL辣根过氧化物酶标记的羊抗人IgG(H+L)二抗(Jackson)工作液,置于37℃孵育30min,用PBST洗板后加入每孔50μL TMB,置于室温避光显色5min后,每孔加入50μL终止液(2M H
2SO
4溶液)终止显色反应。立即把酶标板放入酶标仪中,选择450nm光波长读取酶标板各孔的OD数值。用SoftMax Pro6.2.1软件对数据进行分析处理。以抗体浓度为横坐标,吸光度值为纵坐标进行4-parameter拟合曲线作图,得到抗体的结合EC50值。
检测结果见表1和图6-8所示,人PD-1蛋白五个突变体D29A、E61A、K78A、E84A、L128A与抗PD-1抗体结合EC50分别为0.021nM、2.47×10
12nM、0.023nM、0.022nM、4.207nM。E61A和L128A两个突变体与抗PD-1抗体的结合能力明显降低。
表1 抗PD-1抗体与人PD-1突变体的结合ELISAEC50结果
2.Fortebio Kinetics方法检测抗PD-1抗体14C12H1L1(hG1TM)与人PD-1蛋白突变体的结合活性
首先将抗PD-1抗体用PBS(含0.02%Tween-20,0.1%BSA,pH7.4)稀释至5μg/mL后固定于AHC传感器(Fortebio)表面,时间为120s,传感器在缓冲液中平衡60s,固定在传感器上的抗PD-1抗体与各PD1-mFc突变体结合,浓度为1.24-100nM(三倍稀释),时间120s,蛋白在缓冲液中解离,时间300s。检测温度为37℃,检测频率为0.3Hz,样品板震动速率为1000rpm。数据以1∶1模型拟合分析,得到亲和力常数。
抗PD-1抗体与各PD-1突变体结合结果见图9-14和表2所示,人PD-1蛋白五个突变体D29A、E61A、K78A、E84A、L128A与抗PD-1抗体结合亲和力常数分别为4.25E-10M、3.30E-08M、1.31E-10M、6.65E-10M、7.68E-09M。与结合ELISA结果一致,E61A和L128A两个突变体与抗PD-1抗体的结合能力明显降低。
表2 抗PD-1抗体与人PD-1突变体的Fortebio Kinetics结果
Claims (10)
- 肽,优选结合抗PD-1抗体或其抗原结合片段,所述肽包含结构单元1和结构单元2,其中结构单元1包含选自以下各项组成的组的PD-1蛋白片段:PD-1蛋白的28位-86位的氨基酸(如SEQ ID NO:10所示),28位-85位的氨基酸(如SEQ ID NO:11所示),29位-86位的氨基酸(如SEQ ID NO:12所示),29位-85位的氨基酸(如SEQ ID NO:13所示),58位-86位的氨基酸(如SEQ ID NO:14所示)、58位-85位的氨基酸(如SEQ ID NO:15所示),59位-86位的氨基酸(如SEQI ID NO:16所示)和59位-85位的氨基酸(如SEQ ID NO:17所示),结构单元2包含选自以下各项组成的组的PD-1蛋白片段:PD-1蛋白的127位-133位的氨基酸(如SEQ ID NO:18所示),127位-132位的氨基酸(如SEQ ID NO:19所示),128位-133位的氨基酸(如SEQ ID NO:20所示),和128位-132位的氨基酸(如SEQ ID NO:21所示)。
- 权利要求1所述的肽,所述肽包含结构单元1和结构单元2,其中结构单元1包含PD-1蛋白的29位-85位的氨基酸(如SEQ ID NO:11所示)或58位-85位的氨基酸(如SEQ ID NO:15所示),结构单元2包含PD-1蛋白的128位-132位的氨基酸(如SEQ ID NO:21所示)或130-132位的氨基酸(如SEQ ID NO:3所示),优选结构单元1包含PD-1蛋白的58位-85位的氨基酸(如SEQ ID NO:15所示),结构单元2包含PD-1蛋白的130-132位的氨基酸(如SEQ ID NO:3所示)。
- 权利要求1或2所述的肽,其中PD-1蛋白的氨基酸序列如SEQ ID NO:1所示。
- 权利要求1-3任一项所述的肽,其中结构单元1包含SEQ ID NO:2或SEQ ID NO:10-17任一项所示的序列,结构单元2包含SEQ ID NO:3或SEQ ID NO:18-21任一项所示的序列,优选结构单元1包含SEQ ID NO:2所示的序列,结构单元2包含SEQ ID NO:3所示的序列。
- 权利要求1-4任一项所述的肽,其中所述抗PD-1抗体的抗原结合片段为Fab片段、Fab’、(Fab’) 2、Fab’-SH、Fab/c、Fv、单链抗体(例如,scFv)。
- 权利要求5所述的肽,其中所述Fab片段的重链氨基酸序列如SEQ ID NO:4所示,轻链氨基酸序列如SEQ ID NO:5所示。
- 权利要求1-6任一项所述的肽,其中PD-1蛋白的T59、E61、S62、E84、D85氨基酸与抗PD-1抗体(例如14C12H1L1-Fab)重链的Y32、D33、S52、G54、Y57、Y100位氨基酸结合,PD-1蛋白的P130、K131、A132氨基酸与抗PD-1抗体轻链的F325、E324、D323位氨基酸结合,PD-1蛋白的N58糖基化侧链与抗PD-1抗体(例如14C12H1L1-Fab)重链氨基酸(例如S31、G53、G54和R56)结合。
- 权利要求7所述的肽,其中所述糖基化侧链包含甘露糖,N-乙酰氨基葡萄糖;岩藻糖和β-D-甘露糖。
- 权利要求1-8任一项所述的肽在筛选结合PD-1的抗体或其抗原结合片段中的用途。
- SEQ ID NO:1所示的PD-1蛋白的D29,T59,E61,S62,K78,E84,D85,L128,P130,A132或其组合在筛选结合PD-1的抗体或其抗原结合片段中的用途。
Applications Claiming Priority (2)
| Application Number | Priority Date | Filing Date | Title |
|---|---|---|---|
| CN202011068655 | 2020-09-30 | ||
| CN202011068655.4 | 2020-09-30 |
Publications (1)
| Publication Number | Publication Date |
|---|---|
| WO2022068919A1 true WO2022068919A1 (zh) | 2022-04-07 |
Family
ID=80951242
Family Applications (1)
| Application Number | Title | Priority Date | Filing Date |
|---|---|---|---|
| PCT/CN2021/122062 WO2022068919A1 (zh) | 2020-09-30 | 2021-09-30 | 结合pd-1抗体的肽及其应用 |
Country Status (2)
| Country | Link |
|---|---|
| CN (1) | CN114316022A (zh) |
| WO (1) | WO2022068919A1 (zh) |
Citations (7)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| CN107840887A (zh) * | 2016-09-21 | 2018-03-27 | 基石药业(苏州)有限公司 | 一种新的pd‑1单克隆抗体 |
| US20180118829A1 (en) * | 2016-11-02 | 2018-05-03 | Jounce Therapeutics, Inc. | Antibodies to pd-1 and uses thereof |
| US20190077866A1 (en) * | 2015-12-02 | 2019-03-14 | Stcube, Inc. | Antibodies specific to glycosylated pd-1 and methods of use thereof |
| US20190218297A1 (en) * | 2016-07-20 | 2019-07-18 | Stcube, Inc. | Methods of cancer treatment and therapy using a combination of anitbodies that bind glycosylated pd-l1 |
| US20190270815A1 (en) * | 2015-08-11 | 2019-09-05 | Wuxi Biologics (Cayman) Inc. | Novel anti-pd-1 antibodies |
| US20190322749A1 (en) * | 2017-01-06 | 2019-10-24 | Crescendo Biologics Limited | Single Domain Antibodies to Programmed Cell Death (PD-1) |
| US20200002420A1 (en) * | 2016-09-21 | 2020-01-02 | Cstone Pharmaceuticals | The novel monoclonal antibodies to programmed death 1 (pd-1) |
-
2021
- 2021-09-30 WO PCT/CN2021/122062 patent/WO2022068919A1/zh active Application Filing
- 2021-09-30 CN CN202111157785.XA patent/CN114316022A/zh active Pending
Patent Citations (7)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| US20190270815A1 (en) * | 2015-08-11 | 2019-09-05 | Wuxi Biologics (Cayman) Inc. | Novel anti-pd-1 antibodies |
| US20190077866A1 (en) * | 2015-12-02 | 2019-03-14 | Stcube, Inc. | Antibodies specific to glycosylated pd-1 and methods of use thereof |
| US20190218297A1 (en) * | 2016-07-20 | 2019-07-18 | Stcube, Inc. | Methods of cancer treatment and therapy using a combination of anitbodies that bind glycosylated pd-l1 |
| CN107840887A (zh) * | 2016-09-21 | 2018-03-27 | 基石药业(苏州)有限公司 | 一种新的pd‑1单克隆抗体 |
| US20200002420A1 (en) * | 2016-09-21 | 2020-01-02 | Cstone Pharmaceuticals | The novel monoclonal antibodies to programmed death 1 (pd-1) |
| US20180118829A1 (en) * | 2016-11-02 | 2018-05-03 | Jounce Therapeutics, Inc. | Antibodies to pd-1 and uses thereof |
| US20190322749A1 (en) * | 2017-01-06 | 2019-10-24 | Crescendo Biologics Limited | Single Domain Antibodies to Programmed Cell Death (PD-1) |
Also Published As
| Publication number | Publication date |
|---|---|
| CN114316022A (zh) | 2022-04-12 |
Similar Documents
| Publication | Publication Date | Title |
|---|---|---|
| ES2821753T3 (es) | Procedimientos de producción de Fab y de anticuerpos biespecíficos | |
| CN113943366A (zh) | 新型抗体框架 | |
| JP2023553758A (ja) | 抗SIRPα抗体およびその適用 | |
| DK2504359T3 (da) | Monospecifikke polypeptidreagenser | |
| CN113272651B (zh) | 鉴别蛋白质中的游离巯基的方法 | |
| JP2014515740A (ja) | 抗凝固薬の解毒剤 | |
| García-Ramírez et al. | A native IgE in complex with profilin provides insights into allergen recognition and cross-reactivity | |
| WO2016031932A1 (ja) | アルカリ洗浄によるFc領域を有するタンパク質の製造方法 | |
| CN114761426A (zh) | 通过序列优化生产稳定性增加的生物治疗药物的方法 | |
| WO2022068919A1 (zh) | 结合pd-1抗体的肽及其应用 | |
| Castellana et al. | Resurrection of a clinical antibody: Template proteogenomic de novo proteomic sequencing and reverse engineering of an anti‐lymphotoxin‐α antibody | |
| Hatfield et al. | Specific location of galactosylation in an afucosylated antiviral monoclonal antibody affects its FcγRIIIA binding affinity | |
| Peissert et al. | Selection of a PD‐1 blocking antibody from a novel fully human phage display library | |
| KR20210111767A (ko) | 절단된 다가 다량체 | |
| US20210380719A1 (en) | Rabbit antibodies to human immunoglobulins g | |
| WO2019189940A1 (ja) | 抗原の処理方法。 | |
| US9499633B2 (en) | Dabigatran antidotes | |
| US20220169710A1 (en) | Antibody for enrichment of cells | |
| JP7498747B2 (ja) | 抗gm2ap抗体及びその応用 | |
| US10078082B2 (en) | Detection of free and protein-bound non-human gal-alpha(1-3)-gal epitope | |
| WO2022253156A1 (en) | Monoclonal antibodies against cldn18.2 and fc-engineered versions thereof | |
| WO2024229245A2 (en) | High throughput fab antibody fragment generation and ranking | |
| JP2023073687A (ja) | 改変型抗体 | |
| Koseki et al. | Key role of P230 in the hinge region on the IgG architecture and function | |
| KR101650046B1 (ko) | 영장류 cd19에 특이적인 항체 및 이를 생산하는 융합세포주 |
Legal Events
| Date | Code | Title | Description |
|---|---|---|---|
| 121 | Ep: the epo has been informed by wipo that ep was designated in this application |
Ref document number: 21874573 Country of ref document: EP Kind code of ref document: A1 |
|
| NENP | Non-entry into the national phase |
Ref country code: DE |
|
| 32PN | Ep: public notification in the ep bulletin as address of the adressee cannot be established |
Free format text: NOTING OF LOSS OF RIGHTS PURSUANT TO RULE 112(1) EPC |
|
| 122 | Ep: pct application non-entry in european phase |
Ref document number: 21874573 Country of ref document: EP Kind code of ref document: A1 |