WO2008040734A1 - Genetic cluster of strains of streptococcus thermophilus having appropriate acidifying and texturizing properties for dairy fermentations - Google Patents
Genetic cluster of strains of streptococcus thermophilus having appropriate acidifying and texturizing properties for dairy fermentations Download PDFInfo
- Publication number
- WO2008040734A1 WO2008040734A1 PCT/EP2007/060463 EP2007060463W WO2008040734A1 WO 2008040734 A1 WO2008040734 A1 WO 2008040734A1 EP 2007060463 W EP2007060463 W EP 2007060463W WO 2008040734 A1 WO2008040734 A1 WO 2008040734A1
- Authority
- WO
- WIPO (PCT)
- Prior art keywords
- strain
- strains
- product
- eps
- food
- Prior art date
Links
Classifications
-
- A—HUMAN NECESSITIES
- A23—FOODS OR FOODSTUFFS; TREATMENT THEREOF, NOT COVERED BY OTHER CLASSES
- A23C—DAIRY PRODUCTS, e.g. MILK, BUTTER OR CHEESE; MILK OR CHEESE SUBSTITUTES; MAKING THEREOF
- A23C9/00—Milk preparations; Milk powder or milk powder preparations
- A23C9/12—Fermented milk preparations; Treatment using microorganisms or enzymes
- A23C9/123—Fermented milk preparations; Treatment using microorganisms or enzymes using only microorganisms of the genus lactobacteriaceae; Yoghurt
- A23C9/1238—Fermented milk preparations; Treatment using microorganisms or enzymes using only microorganisms of the genus lactobacteriaceae; Yoghurt using specific L. bulgaricus or S. thermophilus microorganisms; using entrapped or encapsulated yoghurt bacteria; Physical or chemical treatment of L. bulgaricus or S. thermophilus cultures; Fermentation only with L. bulgaricus or only with S. thermophilus
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N1/00—Microorganisms, e.g. protozoa; Compositions thereof; Processes of propagating, maintaining or preserving microorganisms or compositions thereof; Processes of preparing or isolating a composition containing a microorganism; Culture media therefor
- C12N1/20—Bacteria; Culture media therefor
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N1/00—Microorganisms, e.g. protozoa; Compositions thereof; Processes of propagating, maintaining or preserving microorganisms or compositions thereof; Processes of preparing or isolating a composition containing a microorganism; Culture media therefor
- C12N1/20—Bacteria; Culture media therefor
- C12N1/205—Bacterial isolates
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N15/00—Mutation or genetic engineering; DNA or RNA concerning genetic engineering, vectors, e.g. plasmids, or their isolation, preparation or purification; Use of hosts therefor
- C12N15/09—Recombinant DNA-technology
- C12N15/11—DNA or RNA fragments; Modified forms thereof; Non-coding nucleic acids having a biological activity
- C12N15/52—Genes encoding for enzymes or proenzymes
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12R—INDEXING SCHEME ASSOCIATED WITH SUBCLASSES C12C - C12Q, RELATING TO MICROORGANISMS
- C12R2001/00—Microorganisms ; Processes using microorganisms
- C12R2001/01—Bacteria or Actinomycetales ; using bacteria or Actinomycetales
- C12R2001/46—Streptococcus ; Enterococcus; Lactococcus
Definitions
- the invention relates to a genetic cluster of strains of Streptococcus thermophilits (S. therniophilus) having appropriate acidifying and texturi/ing properties for dairy fermentations.
- Bacteriophages are viruses capable of attacking bacteria. During these viral attacks, the bacteriophages infect the bacterial culture, and multiply in order to finally destroy this culture.
- the technological impact on the milk processing industry (production of cheese, yogurt, fermented milk) of these bacteriophages is significant since they can cause a complete cessation of fermentation and therefore prevent the production of these milk-derived products.
- One way of combating the problems linked to the infection of the fermentations by bacteriophages is the use of strains having appropriate sensitivity spectra to the phages.
- producers of dairy ferments have developed strategies to combat the bacteriophages by constructing ferments constituted by several strains having distinct lysotypes. and using several of these ferments in rotation.
- S. therniophilus are used extensively alone or in combination with other bacteria for the production of fermented food products. They are included in particular in the formulation of the ferments used for producing yogurts. Strains of S. thermophilic are expected to participate in the formation of lactic curd by acidification of milk and in the development of the texture of the fermented product. ⁇ distinction is generally drawn between 4 groups of S.
- a texturi/ing strain is a strain making it possible to obtain fermented milks the gels of which can be described by their rheoiogical properties.
- the invention relates to the strains of S. thermophilics in a genetic cluster which have a lysotype distinct from that of the acidifying and texturizing strains of S. thermophilic currently used. Within this cluster novel acidifying and texturizing strains have been identified.
- the invention also describes a method making it possible to predict a strain's membership of a family of strains having identical or related lysotypes. This method analyzes the restriction polymorphism of the epsA-B-C-D region of the genome of S. the ⁇ nophilus.
- the eps ⁇ -B-C-D region is meant the region of the chromosome of S. the ⁇ nophilus overlapping the epsA to epsD genes of the eps locus.
- the DNA fragment corresponding to this region can be obtained by PCR reaction on the chromosomic DNA of S. thermophilic using the oligonucleotides of SEQ ID N° l and SHQ ID N°2 as primers.
- a subject of the present invention is a strain of Streptococcus thermophilus the epsAD fragment of which, after digestion by the restriction enzymes MnIl, Fold and ///HdIII, has a restriction profile characterized by DNA fragments of base pairs (bp), 341 ⁇ 2 bp, 305 ⁇ 2 bp, 299 ⁇ 2 bp, 277 ⁇ 2 bp, 2 I 0 ⁇ 2 bp, 160 ⁇ 2 bp. 142 ⁇ 2 bp. 100 ⁇ 2 bp, 79 ⁇ 2 bp, 75 ⁇ 2 bp, 66 ⁇ 2 bp, 42 ⁇ 2 bp, 23 ⁇ 2 bp and 9 ⁇ 2 bp.
- the restriction profile of the epsAD fragment is determined by Standard sequencing of the epsAD fragment followed by /// silico determination of the restriction profile.
- sequencing can be carried out with the CEQ8000 equipment (Bcckman) and the in silico determination of the restriction profile can be carried out starting from the sequence of the epsAD fragment using the NEBcutter V2.0 tool accessible on the internet via the website https://tools.neb.com/.
- a strain according to the invention comprises a nucleotide sequence having at least 80%, preferentially at least 90%, at least 95% or at least 97% and still more preferentially 100% identity with the nucleotide sequence of SEQ ID NO: 1
- a strain according to the invention comprises a nucleotide sequence having at least 80%, preferentially at least 90%, at least 95% or at least 97% and still more preferentially 100% identity with the nucleotide sequence of SEQ ID NO: 1
- the strain according to the invention is textuiizing.
- the strain according to the invention acidifies rapidly. Still more preferentially the strain according to the invention is textuiizing and acidifies rapidly.
- strain of Streptococcus thermophihts is meant a strain which produces fermented milks having, under the conditions described in the example, a viscosity greater than approximately 35 Pa. s, a thixotropic area of less than approximately 2000 Pa/s and/or a yield point of less than approximately 14 Pa.
- rapidly acidifying strain is meant a strain which under the conditions described in the example, has a Vm of less than -0.0100 upH/min.
- a strain according to the invention is the strain of Streptococcus thennophiliis deposited on 14th June 2006 at the Collection Nationale de Cultures de Microorganisni.es under no. CNCM 1-3617 or a mutant strain which can be obtained from the latter.
- a person skilled in the art can use the usual mutagenesis techniques such as UV irradiation or exposure to mutagenic chemical products (cthyl-methane-sulphonate, nitrosoguanidine, nitrous acid etc.).
- a subject of the invention is the strain of Streptococcus thennophiliis deposited on 14th June 2006 in the name of Danisco France SAS, 20 rue de Brunei, 75017 Paris at the Collection Nationale de Cultures de
- a person skilled in the art starting from the restriction profiles described previously and/or from the sequences of SEQ ID N°4 and/or N°5, can identify the strains which belong to the same genetic cluster as the strain CNCM 1-3617.
- a subject of the invention is also a bacterial composition comprising at least one strain according to the invention.
- bacterial composition is meant a mixture of different strains, in particular a ferment, or a leaven.
- the mixtures of preferred strains according to the invention are mixtures of Streptococcus thennophilus with other Streptococcus thennophilus, or mixtures of Streptococcus thennophilus with Lactobacillus delhriieckii subsp. hulgaricus, or mixtures of Streptococcus thennophilus with other Lactobacillus and/or with Bifidobacterium, or mixtures of Streptococcus thennophilus with Lactococcus. or mixtures of Streptococcus thennophilus with other strains of lactic bacteria and/or yeasts.
- a subject of the invention is also a manufacturing process for a food product, a food complement, a dietary supplement or a product with probiotic properties, comprising a stage in which a strain according to the invention is used.
- the food product, the food complement, the dietary supplement or the product with probiotic properties is a dairy product, a meat product, a cereal product, a drink, a foam or a powder.
- the food product, the food complement, the dietary supplement or the product with probiotic properties is a dairy product. It is for example a fermented milk, a yogurt, a matured cream, a cheese, a fromage frais, a milk drink, a dairy product retentate, a processed cheese, a cream dessert, a cottage cheese or an infant milk.
- the dairy product comprises milk of animal and/or plant origin.
- a subject of the invention is also a food product, a food complement, a dietary supplement or a product with probiotic properties comprising at least one strain according to the invention or the bacterial composition described previously.
- the invention also describes a method for predicting the lysotype of a strain of S. thennophilus starting from analysis of the restriction polymorphism of the epsA- B-C-D region of its genome, comprising the following stages: a) amplification of the epsAD fragment by PCR reaction on the chromosomic DNA of 5.
- strains of S. thermophilus the Iysotype of which is know n are listed in Table 3.
- Figure 1 Diagrammatic representation of the eps locus of various S. thermophilus having similarities in organization and sequence of the 5' region including the eps A, epsB, epsC and eps D genes. The GENBANK access numbers of the nucleotide sequences are given in parentheses.
- Figure 2 Alignment of partial sequences of the epsA (A) and epsD (B) genes at the level of the regions targeted by the primers of SEQ ID N°l (in the epsA gene) and SEQ ID N°2 (in the epsD gene).
- Figure 3 In silico determination of the profiles obtained according to the epsAD method of the various strains of S. thermophilic the sequence of the eps locus of which is described in the literature.
- the GENBANK access numbers of these strains are: ⁇ F448502 (MTC310), AF373595 (Sfi39), AF410175 (Type I).
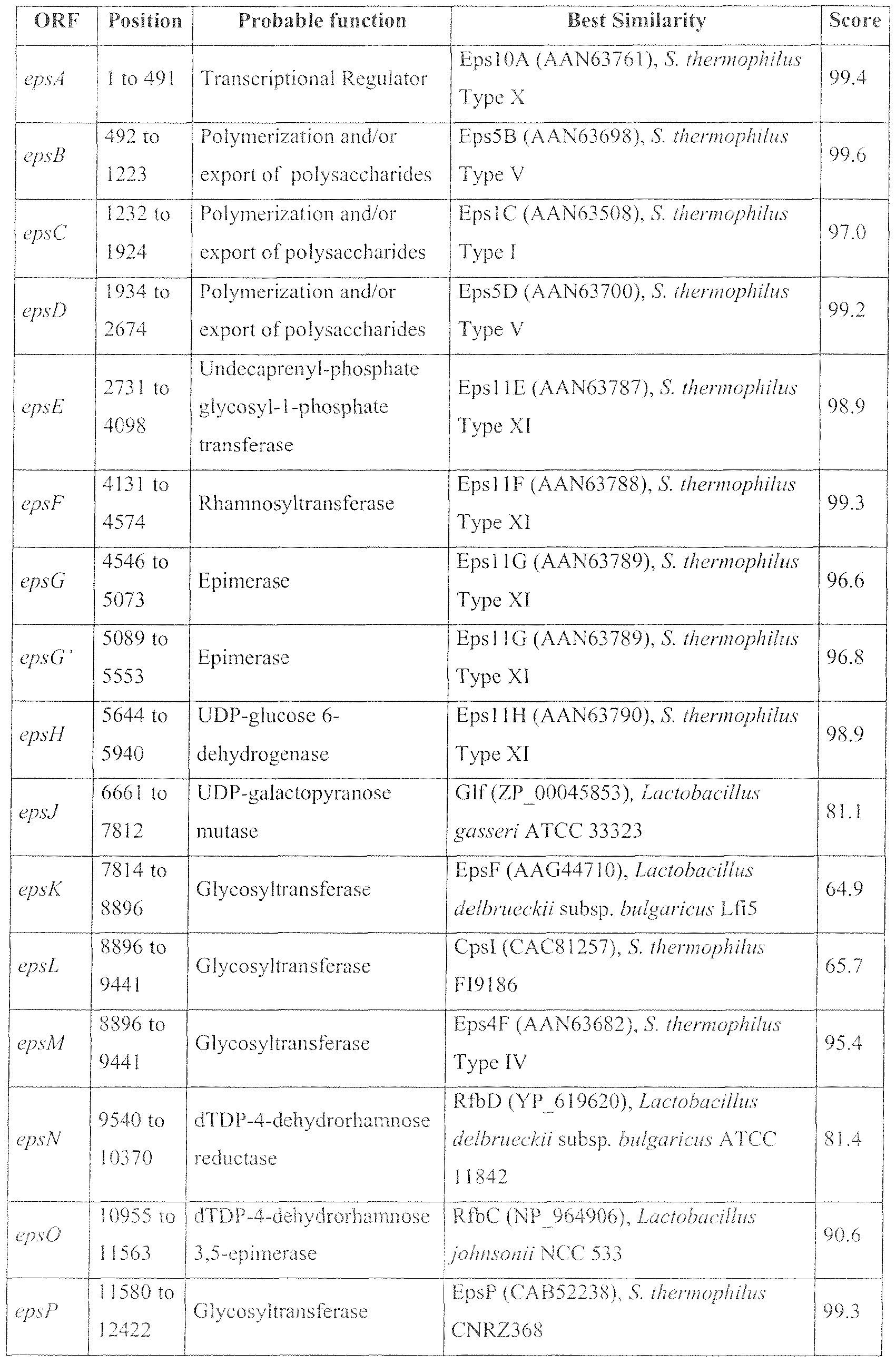
- FIG. 1 Diagrammatic representation of the organization of the eps locus of the strain CNCM 1-3617, corresponding to the sequence SEQ ID N°3.
- the open reading frames (ORF) represented by vertical hatching (region 1 ) ha ⁇ e significant similarities with the epsA-epsB-epsC-epsD genes situated at the start of the eps locus in the great majority of the strains of S. thermophilic
- the ORFs represented by horizontal hatching (region 2) have significant similarities with the eps 1 IE, eps J IF.
- the ORFs represented by a chequered pattern have significant similarities with the eps4F gene of the type IV strain.
- the grey rectangle between regions 2 and 3 represents a non-coding region having sequence homologies with a region of the eps locus of the type IV strain.
- the ORFs represented by diagonal hatching (region 6) have significant similarities with the eps P, epsQ, epsS, epsR and eps T genes of the CNRZ368 strain.
- the unshaded ORFs regions 3 and 5 have no significant similarities with the eps genes described for other S. thermophilic.
- the two black bands at the bottom of the figure represent the position of the sequences SEQ ID N°4 and SEQ ID N°5.
- Table 1 and Table 3 show some of the strains used for the study. Some of these strains are obtained from the Danisco collection of strains and phages (DGCC: Danisco Global Culture Collection). The preparation of cultures of these strains was carried out according to the standard methods of microbiology.
- DGCC Danisco Global Culture Collection
- a on the basis of industrial use and the present study.
- b on the basis of published sequence results of the eps locus and/or the structure of the polysaccharide and data internal to Danisco.
- c on the strain studied and/or on related strains.
- the strain CNCM 1-3617 belongs to a novel genetic cluster
- the recent determination of the complete sequence of the chromosome of two strains of S. thermophilic CNRZ 1066 and LMG 183 1 1 shows a high level of conservation of the genetic content and the organization of the genes in this species) Bolotin et al., 2004).
- One of the rare regions exhibiting major genetic differences between these two strains corresponds Io the eps locus which codes for the genes involved in the biosynthesis of exopolysaccharides.
- a great diversity has already been described at the level of this genetic locus (cf. Figure 1 ) since several eps sequences had been determined for various strains of S.
- thermophilus (for a journal article, see Broadbcnt et al., 2003). Flanked by the deoD (coding for a purine-nucleoside phosphorylase) and pgm (coding for a phospho-glucomutase) genes, all the clusters of eps genes in S. thermophilus are composed of a conserved proximal region (from the epsA gene to the epsD gene) and of a highly variable distal region. In spite of a conserved organization of the epsA-B-C-D genes, a significant sequence polymorphism exists in this region. The polymorphism of this region has been used to develop a tool for genetic typing of the strains of S. thermophilus: the epsAD method.
- epsAD method The tool developed is based on the specific amplification (PCR) of the epsA-B-C-D region followed by the analysis of its restriction polymorphism
- composition of the reaction mixture (50 ⁇ L): buffer for the DNA polymerase ⁇ l ,
- the PCR product is checked by electrophoresis on 1.5% agarose gel.
- the size of the amplified product is approximately 2480 bp.
- the sequence of the PCR fragment is determined (according to a method derived from Sanger et al., 1977) with CEQ8000 equipment (Beckman).
- the sequence is processed by the NEBcutter tool (for example, https://tools.neb.com/) by selecting the restriction enzymes Mull, Fokl and HmdIII in order to establish its restriction profile in silico, and in particular in order to define the size of the restriction fragments.
- Table 2 Size of the DNA fragments determined by in silico digestion of the epsAD fragment by the restriction enzymes MnIl, Fokl and HindlU, for the strains of S. thermophilic the sequence of the epsA-B-C-D genes of which is available.
- MTC310 AF448502 778; 371 ; 344; 299; 175; 142; 100; 79; 75; 42; 35; 23; 9
- the PCR product can be digested by the restriction enzymes MnIl, Fokl and H/V/dIII under the following conditions: PCR product 15 to 30 ⁇ L. buffer 2 (New England Biolabs) ⁇ l , bovine serum albumin (New England Biolabs) ⁇ l , enzyme MnIl (New England Biolabs) 1 unit, enzyme Fokl (New England Biolabs) 1 unit, enzyme H///dIII (New England Biolabs) 1 unit, H 2 O qsf 50 ⁇ L. Incubation at 37°C for 1 hour.
- the restriction fragments are then analyzed by electrophoresis. Electrophoresis on agarose gel can be used. However, in order to remedy the low resolution power of this type of electrophoresis (precision +/- 10%) and the difficulty of visualizing fragments smaller than 100 bp, methods with a higher resolution (+/- 0.1 to 1%) such as micro-fiuidic electrophoresis (Agilent) or capillary electrophoresis may be preferred.
- Tableau 3 Summary of the results of genotyping and lysotyping
- Method for determining the sensitivity of a strain to a bacteriophage The sensitivity of a strain to a bacteriophage is established by the lysis plaque method. 100 ⁇ l of a culture of the strain to be tested and 100 ⁇ l of an appropriate dilution of a serum containing the bacteriophage to be studied are used in order to seed 5 ml of an agar medium under surfusion (0.6 % agar weight/volume) M 17i-glucosc supplemented at 10 mM with CaCb. The mixture is poured onto the surface of a solidified agar medium ( 1.5 % agar weight/volume) M 17+glucosc supplemented at 10 mM with CaCl 2 .
- strain's sensitivity to the bacteriophage is evaluated by the presence of lysis plaques.
- the absence of lysis plaque signifies this strain's resistance to the bacteriophages tested.
- the spectrum of a strain's sensitivity to the bacteriophages, also called lysotypc is constituted by all of the sensitivities and resistances to the bacteriophages studied.
- a reference system with approximately sixty phages has been implemented in order to establish the lysotype of the strains of S. thermophilus in the Danisco collection.
- the strains which have the same lysotype have been grouped together in different groups denoted LT-n.
- the strain CNCM 1-3617 belongs to a novel genetic group called CL-I which has the very particular lysotype of being resistant to all the bacteriophages tested.
- thermophilus has shown that the eps locus is one of the major sites of heterogeneity between strains. This characteristic has already been exploited in part in order to develop the abovementioned genotyping method which uses the diversity in the epsA-B-C-D region which is the proximal region of the eps locus. An even greater diversity appears in the distal region of the eps locus (region encoding the glycosyl- transferases, see Figure 1 ). This region gives the specificity of the exopolysaccharide and therefore induces at least in part the specificity of the strain's thickening power.
- the nucleotide sequence of the eps locus of the strain CNCM 1-3617 (starting from the epsA gene) was obtained from a synthetic DNA fragment. This fragment was synthesized by PCR on a purified genomic DNA matrix of the strain CNCM 1-3617 using two specific primers of conserved genes (deal) encoding a purine-nucleotide phosphorylase, and orfl4.9 of unknown function) generally framing the eps locus in the S. thermophilus described in the literature.
- sequence of 16037 bp originating from the strain CNCM 1-3617 corresponds to the SEQ ID N C 3.
- the sequences SEQ ID N°4 and SEQ ID N°5 correspond respectively to the positions 6592 to 9391 and 10331 to 1 1373 of SEQ ID N°3.
- FIG. 4 shows diagranimatically the genetic structure of the eps locus of the strain CNCM 1-3617 established by analysis of its nucleotide sequence.
- the part upstream of the epsA open reading frame (QRF), the sequence of the start of the epsA ORF, and the part downstream of the epsT QRb are not known.
- Analysis of the sequence identifies 20 ORFs which are all oriented in the same direction. Due to their similarity of sequence with other known genes, and/or by the presence of specific protein units within products deduced from these QRFs, it is possible to attribute a putative function to them.
- Region 1 this region is formed from 4 ORFs ⁇ eps A, epsB, epsC, eps D) for which the deduced proteins exhibit very great similarities (between 95.7 and 99.6%) with the proteins deduced from ORFs situated in the eps locus of strains of S. therinophilus.
- Region 2 this region is formed from 5 ORFs (epsE, eps F, epsG, epsC and epslf) for which the deduced proteins exhibit very great similarities (between 96.6 and 99.3%) with the proteins deduced from ORFs situated in the eps locus of the type XI strain (GENBANK access no. AF454501 ).
- Region 3 this region is formed from 3 ORFs ⁇ eps J. cpsK and epsL) for which the deduced proteins exhibit sufficient similarities (less than 81 %) with proteins described in the literature to assign a probable function to them. However these ORFs are clearly distinct from ORFs already described in the literature.
- Region 4 this region is formed from one ORF (epsM) for which the deduced protein exhibits very great similarities (95.4%) with the protein deduced from the eps4F ORF situated in the eps locus of the type IV strain.
- Region 5 this region is formed from 2 ORFs (epsN and epsO) for which the deduced proteins exhibit sufficient similarities (less than 91 %) with proteins of lactobacillae described in the literature to assign a probable function to them.
- these ORFs are clearly distinct from ORFs already described in the literature in S. thermophilic
- Region 6 this region is formed from 5 ORFs (epsP, epsO, epsS, eps R and eps T) for which the deduced proteins exhibit very great similarities
- Table 4 Analysis of the ORFs of the eps locus of the strain CNCM 1-3617.
- the position corresponds to the nucleotides at the start and end of the ORF in the sequence SEQ ID N°3.
- the probable function is that of the proteins deduced from the ORF sequence.
- the best similarity is the protein or the deduced protein (the GENBANK access number of the protein sequence is indicated in parentheses) having the greatest similarity with the protein deduced from the ORF.
- the score is the percentage similarity obtained for the best similarity using the "blastp" tool (Protein-protein BLAST, https://www.ncbi.nlm.nih.gov/BLAST/).
- the fermentation support is obtained by supplementing 100 nil of semi-skimmed UHT milk (Le Petit Vendeen®) with 3% (weight/volume) of skimmed milk powder (SUP ' R TOP®, Eurial Poitouraine).
- the sterility of the solution is obtained by pasteurization for 10 min at 90 0 C (at the core).
- the fermentation support thus obtained is inoculated with the strain to be tested at a rate of 10 1 cfu/ml, then incubated at 43°C (in a water bath).
- the pH is continuously monitored using a CINAC apparatus (Ysebaert).
- thermophihis can be described by the maximum rate of acidification, Vm (pH unit/min (pHu/min)), calculated by the maximum value of the first derivative of the pH curve as a function of time. Under these operating conditions, it is estimated that this variable is characteristic of the strain. Its value is constant whatever the physiological state of the microorganism upon inoculation of the milk and the level of seeding. Two groups of strains are distinguished, the strains with so-called slow acidification, the Vm of which is greater than - 0.0100 pHu/min, and the strains with so-called rapid acidification the Vm of which is les than -0.0100 pHu/min.
- the strain CNCM I- 3617 clearly belongs to the group of the so-called rapid acidification strains (Table 5). This propeity i.s ⁇ e ⁇ y often linked to the presence in the genome of the strains of a gene encoding the wall protease PrtS which could be detected in the genome of CNCM 1-3617.
- Table 5 Maximum iatc of acidification of different stiams of Sticptococcus the) mophihts e ⁇ sin under the opciating conditions desci ibcd
- the lei mentation suppoit is obtained by supplementing 100 ml of semi-skimmed LHT milk (Le Petit Vendecn®) with 3% (weight/volume) ot bkimmed milk powder (SUP'R TOP ⁇ , Eu ⁇ al Poitouraine)
- the sterility of the solution is obtained by pasteurization for 10 min at 9O 0 C (at the core)
- the fermentation support thus obtained is inoculated with the strain to be tested at a rate of 10 ( cfu'ml, then incubated at 43°C (in a w atei bath) until a pH of 4 6 is obtained
- the pH is continuously monitored using a CINAC apparatus (Ysebaeit)
- the lamented milks thus obtained are placed in a ⁇ entilattd oven at 6°C until they aie analyzed Tw o types of theological measurements aie carried out ⁇ iscosity and flow
- the ⁇ iscosity measurements are cai ⁇
- the flow measurements arc cariied out at a temperature of 8°C on previously-stirred fermented milks, after storage for 14 days at 6 0 C,
- the stress applied in a continuous sweep varies from 0 to 60 Pa for a duration of 1 min according to a linear mode.
- the stress applied in a continuous sweep varies from 60 to 0 Pa for a duration of 1 min according to a linear mode.
- the values taken into account are the thixotropic area and the yield point; the
- I u latter is calculated according to the Casson model.
- the texturizing ability of a strain can be evaluated in a first phase by a viscosity measurement of the curd obtained under the operating conditions described above.
- the recognized non-texturizing strains provide viscosity values close to 30 Pa. s while the texturizing strains exceed 40 Pa. s. This texturizing ability can be more
- strain CNCM 1-2979 produces a curd the viscosity of which reaches 42 Pa. s, and the strain DGCC7966 makes it possible to obtain a clearly higher viscosity, of 70 Pa.s.
- the strain CNCM 1-3617 provides curds the viscosity of which amounts to 54 Pa.s (Table 6). This value places this strain among the group of strains with a texturizing ability fully 0 comparable to the industrial strains currently used to devise lactic ferments for the production of yogurts and fermented milks.
- Tableau 7 Values of yield point and thixotropic area, Casson model, measurements by the ARl OOO-N on fermented dairy products with different strains after storage for 14 days at 6°C.
- the strain CNCM 1-3617 has several characteristics of interest for the construction of ferments and in particular for ferments used during the production of yogurts or fermented milks. It exhibits a rare combination of functional properties (acidifying and thickening strain) and its lysotype is distinct from that of the other strains used in a standard manner for these applications.
- Streptococcus thermophilus a review.
- BLAST 2 Sequences a new tool for comparing protein and nucleotide sequences.
Landscapes
- Life Sciences & Earth Sciences (AREA)
- Engineering & Computer Science (AREA)
- Health & Medical Sciences (AREA)
- Genetics & Genomics (AREA)
- Chemical & Material Sciences (AREA)
- Biotechnology (AREA)
- Organic Chemistry (AREA)
- Zoology (AREA)
- Bioinformatics & Cheminformatics (AREA)
- Wood Science & Technology (AREA)
- Biomedical Technology (AREA)
- Microbiology (AREA)
- General Engineering & Computer Science (AREA)
- General Health & Medical Sciences (AREA)
- Biochemistry (AREA)
- Virology (AREA)
- Tropical Medicine & Parasitology (AREA)
- Medicinal Chemistry (AREA)
- Molecular Biology (AREA)
- Physics & Mathematics (AREA)
- Biophysics (AREA)
- Plant Pathology (AREA)
- Food Science & Technology (AREA)
- Polymers & Plastics (AREA)
- Measuring Or Testing Involving Enzymes Or Micro-Organisms (AREA)
- Micro-Organisms Or Cultivation Processes Thereof (AREA)
- Dairy Products (AREA)
- Enzymes And Modification Thereof (AREA)
Abstract
Description
Claims
Priority Applications (7)
| Application Number | Priority Date | Filing Date | Title |
|---|---|---|---|
| EP07820845A EP2076584B1 (en) | 2006-10-03 | 2007-10-02 | Genetic cluster of strains of streptococcus thermophilus having appropriate acidifying and texturizing properties for dairy fermentations |
| DK07820845.1T DK2076584T3 (en) | 2006-10-03 | 2007-10-02 | Genetic cluster of strains of Streptococcus thermophilus with suitable acidifying and texturing properties for milk fermentation |
| MX2009003651A MX2009003651A (en) | 2006-10-03 | 2007-10-02 | Genetic cluster of strains of streptococcus thermophilus having appropriate acidifying and texturizing properties for dairy fermentations. |
| BRPI0717777-1A2A BRPI0717777A2 (en) | 2006-10-03 | 2007-10-02 | GENETIC GROUPPING OF STRETOCOCCUS THERMOPHILUS CEPAS HAVING APPROPRIATE ACIDIFICATION AND TEXTURIZATION PROPERTIES FOR MILK FERMENTATIONS |
| US12/443,903 US8236550B2 (en) | 2006-10-03 | 2007-10-02 | Genetic cluster of strains of Streptococcus thermophilus having appropriate acidifying and texturizing properties for dairy fermentations |
| PL07820845T PL2076584T3 (en) | 2006-10-03 | 2007-10-02 | Genetic cluster of strains of streptococcus thermophilus having appropriate acidifying and texturizing properties for dairy fermentations |
| CN2007800417747A CN101595210B (en) | 2006-10-03 | 2007-10-02 | Genetic cluster of strains of streptococcus thermophilus having appropriate acidifying and texturizing properties for dairy fermentations |
Applications Claiming Priority (2)
| Application Number | Priority Date | Filing Date | Title |
|---|---|---|---|
| FR0608657A FR2906536B1 (en) | 2006-10-03 | 2006-10-03 | GENETIC CLUSTER OF STREPTOCOCCUS THERMOPHILUS STRAINS HAVING ACIDIFTING AND TEXTURANT PROPERTIES SUITABLE FOR DAIRY FERMENTATION. |
| FR0608657 | 2006-10-03 |
Publications (2)
| Publication Number | Publication Date |
|---|---|
| WO2008040734A1 true WO2008040734A1 (en) | 2008-04-10 |
| WO2008040734A9 WO2008040734A9 (en) | 2009-09-11 |
Family
ID=37775534
Family Applications (1)
| Application Number | Title | Priority Date | Filing Date |
|---|---|---|---|
| PCT/EP2007/060463 WO2008040734A1 (en) | 2006-10-03 | 2007-10-02 | Genetic cluster of strains of streptococcus thermophilus having appropriate acidifying and texturizing properties for dairy fermentations |
Country Status (13)
| Country | Link |
|---|---|
| US (1) | US8236550B2 (en) |
| EP (1) | EP2076584B1 (en) |
| CN (1) | CN101595210B (en) |
| AR (1) | AR063029A1 (en) |
| BR (1) | BRPI0717777A2 (en) |
| CL (1) | CL2007002801A1 (en) |
| DK (1) | DK2076584T3 (en) |
| FR (1) | FR2906536B1 (en) |
| MX (1) | MX2009003651A (en) |
| PL (1) | PL2076584T3 (en) |
| RU (1) | RU2009116438A (en) |
| TW (1) | TW200823288A (en) |
| WO (1) | WO2008040734A1 (en) |
Cited By (9)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| WO2011004012A2 (en) | 2009-07-10 | 2011-01-13 | Chr. Hansen A/S | Production of cottage cheese by using streptococcus thermophilus |
| WO2011026863A1 (en) | 2009-09-01 | 2011-03-10 | Chr. Hansen A/S | Lactic bacterium with modified galactokinase expression for texturizing food products by overexpression of exopolysaccharide |
| WO2011092300A1 (en) | 2010-01-28 | 2011-08-04 | Chr. Hansen A/S | Lactic bacterium for texturizing food products selected on basis of phage resistance |
| WO2012145629A2 (en) | 2011-04-20 | 2012-10-26 | Danisco A/S | Production of cheese with s. thermophilus |
| JP2012531189A (en) * | 2009-06-30 | 2012-12-10 | セーホーエル.ハンセン アクティーゼルスカブ | Method for the production of fermented dairy products |
| US8771766B2 (en) | 2008-12-12 | 2014-07-08 | Dupont Nutrition Biosciences Aps | Genetic cluster of strains of Streptococcus thermophilus having unique rheological properties for dairy fermentation |
| US9453231B2 (en) | 2010-10-22 | 2016-09-27 | Chr. Hansen A/S | Texturizing lactic acid bacteria strains |
| WO2018114573A1 (en) * | 2016-12-20 | 2018-06-28 | Dsm Ip Assets B.V. | Streptococcus thermophilus starter cultures |
| WO2018220104A1 (en) * | 2017-05-31 | 2018-12-06 | Chr. Hansen A/S | Method of producing streptococcus thermophilus mutant strains |
Families Citing this family (3)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| US9635870B2 (en) | 2011-02-28 | 2017-05-02 | Franklin Foods Holdings Inc. | Direct-set cheese |
| US9462817B2 (en) | 2011-02-28 | 2016-10-11 | Franklin Foods Holdings Inc. | Processes for making cheese products utilizing denatured acid whey proteins |
| CN112280795A (en) * | 2020-11-17 | 2021-01-29 | 昆明理工大学 | Use of glycosyltransferase genes |
Citations (1)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| WO2004085607A2 (en) * | 2003-03-17 | 2004-10-07 | Danisco France | Texturizing lactic bacteria |
Family Cites Families (2)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| FR2723963B1 (en) * | 1994-08-31 | 1997-01-17 | Gervais Danone Co | PREPARATION OF PRODUCTS FERMENTED BY STREPTOCOCCUS THERMOPHILUS, ENRICHED IN GALACTO-OLIGOSACCHARIDES AND BETA-GALACTOSIDASE |
| CN1793325A (en) * | 2005-11-30 | 2006-06-28 | 东北农业大学 | Aromatic type direct putting type ferment agent for sour milk |
-
2006
- 2006-10-03 FR FR0608657A patent/FR2906536B1/en not_active Expired - Fee Related
-
2007
- 2007-09-27 AR ARP070104281A patent/AR063029A1/en unknown
- 2007-09-28 CL CL200702801A patent/CL2007002801A1/en unknown
- 2007-10-02 CN CN2007800417747A patent/CN101595210B/en not_active Expired - Fee Related
- 2007-10-02 EP EP07820845A patent/EP2076584B1/en not_active Not-in-force
- 2007-10-02 PL PL07820845T patent/PL2076584T3/en unknown
- 2007-10-02 BR BRPI0717777-1A2A patent/BRPI0717777A2/en not_active IP Right Cessation
- 2007-10-02 WO PCT/EP2007/060463 patent/WO2008040734A1/en active Application Filing
- 2007-10-02 RU RU2009116438/10A patent/RU2009116438A/en not_active Application Discontinuation
- 2007-10-02 MX MX2009003651A patent/MX2009003651A/en not_active Application Discontinuation
- 2007-10-02 US US12/443,903 patent/US8236550B2/en active Active
- 2007-10-02 DK DK07820845.1T patent/DK2076584T3/en active
- 2007-10-03 TW TW096137159A patent/TW200823288A/en unknown
Patent Citations (1)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| WO2004085607A2 (en) * | 2003-03-17 | 2004-10-07 | Danisco France | Texturizing lactic bacteria |
Non-Patent Citations (2)
| Title |
|---|
| BROADBENT J R ET AL: "Biochemistry, genetics, and applications of exopolysaccharide production in Streptococcus thermophilus: a review", JOURNAL OF DAIRY SCIENCE, AMERICAN DAIRY SCIENCE ASSOCIATION, SAVOY, IL, US, vol. 86, no. 2, February 2003 (2003-02-01), pages 407 - 423, XP009018436, ISSN: 0022-0302 * |
| JOLLY LAURE ET AL: "Molecular organization and functionality of exopolysaccharide gene clusters in lactic acid bacteria", INTERNATIONAL DAIRY JOURNAL, vol. 11, no. 9, 2001, pages 733 - 745, XP002423540, ISSN: 0958-6946 * |
Cited By (23)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| US9149049B2 (en) | 2008-12-12 | 2015-10-06 | Dupont Nutrition Biosciences Aps | Genetic cluster of strains of streptococcus thermophilus having unique rheological properties for dairy fermentation |
| EP2789687A1 (en) | 2008-12-12 | 2014-10-15 | DuPont Nutrition Biosciences ApS | Genetic cluster of strains of Streptococcus thermophilus having unique rheological properties for dairy fermentation |
| US8771766B2 (en) | 2008-12-12 | 2014-07-08 | Dupont Nutrition Biosciences Aps | Genetic cluster of strains of Streptococcus thermophilus having unique rheological properties for dairy fermentation |
| JP2012531189A (en) * | 2009-06-30 | 2012-12-10 | セーホーエル.ハンセン アクティーゼルスカブ | Method for the production of fermented dairy products |
| US9028896B2 (en) | 2009-07-10 | 2015-05-12 | Chr. Hansen A/S | Production of cottage cheese by using Streptococcus thermophilus |
| WO2011004012A2 (en) | 2009-07-10 | 2011-01-13 | Chr. Hansen A/S | Production of cottage cheese by using streptococcus thermophilus |
| EA020272B1 (en) * | 2009-07-10 | 2014-09-30 | Кр. Хансен А/С | Production of cottage cheese by using streptococcus thermophilus |
| WO2011004012A3 (en) * | 2009-07-10 | 2011-07-21 | Chr. Hansen A/S | Production of cottage cheese by using streptococcus thermophilus |
| WO2011026863A1 (en) | 2009-09-01 | 2011-03-10 | Chr. Hansen A/S | Lactic bacterium with modified galactokinase expression for texturizing food products by overexpression of exopolysaccharide |
| US9416351B2 (en) | 2010-01-28 | 2016-08-16 | Chr. Hansen A/S | Lactic bacterium for texturizing food products selected on basis of phage resistance |
| US9060524B2 (en) | 2010-01-28 | 2015-06-23 | Chr. Hansen A/S | Bacterium |
| WO2011092300A1 (en) | 2010-01-28 | 2011-08-04 | Chr. Hansen A/S | Lactic bacterium for texturizing food products selected on basis of phage resistance |
| US9562221B2 (en) | 2010-01-28 | 2017-02-07 | Chr. Hansen A/S | Lactic bacterium for texturizing food products selected on the basis of phage resistance |
| US8865238B2 (en) | 2010-01-28 | 2014-10-21 | Chr. Hansen A/S | Bacterium |
| US10392597B2 (en) | 2010-10-22 | 2019-08-27 | Chr. Hanse A/S | Texturizing lactic acid bacteria strains |
| US9453231B2 (en) | 2010-10-22 | 2016-09-27 | Chr. Hansen A/S | Texturizing lactic acid bacteria strains |
| WO2012145629A2 (en) | 2011-04-20 | 2012-10-26 | Danisco A/S | Production of cheese with s. thermophilus |
| WO2018114573A1 (en) * | 2016-12-20 | 2018-06-28 | Dsm Ip Assets B.V. | Streptococcus thermophilus starter cultures |
| US11674117B2 (en) | 2016-12-20 | 2023-06-13 | Dsm Ip Assets B.V. | Streptococcus thermophilus starter cultures |
| AU2017381338B2 (en) * | 2016-12-20 | 2024-02-15 | Dsm Ip Assets B.V. | Streptococcus thermophilus starter cultures |
| WO2018220104A1 (en) * | 2017-05-31 | 2018-12-06 | Chr. Hansen A/S | Method of producing streptococcus thermophilus mutant strains |
| US11564399B2 (en) | 2017-05-31 | 2023-01-31 | Chr. Hansen A/S | Method of producing Streptococcus thermophilus mutant strains |
| US11758914B2 (en) | 2017-05-31 | 2023-09-19 | Chr. Hansen A/S | Method of producing Streptococcus thermophilus mutant strains |
Also Published As
| Publication number | Publication date |
|---|---|
| DK2076584T3 (en) | 2012-07-23 |
| WO2008040734A9 (en) | 2009-09-11 |
| FR2906536B1 (en) | 2008-12-26 |
| PL2076584T3 (en) | 2012-12-31 |
| MX2009003651A (en) | 2009-05-28 |
| US20100184181A1 (en) | 2010-07-22 |
| BRPI0717777A2 (en) | 2014-04-22 |
| RU2009116438A (en) | 2010-11-10 |
| EP2076584A1 (en) | 2009-07-08 |
| CL2007002801A1 (en) | 2008-02-15 |
| CN101595210B (en) | 2013-09-11 |
| TW200823288A (en) | 2008-06-01 |
| EP2076584B1 (en) | 2012-05-30 |
| AR063029A1 (en) | 2008-12-23 |
| US8236550B2 (en) | 2012-08-07 |
| FR2906536A1 (en) | 2008-04-04 |
| CN101595210A (en) | 2009-12-02 |
Similar Documents
| Publication | Publication Date | Title |
|---|---|---|
| EP2076584B1 (en) | Genetic cluster of strains of streptococcus thermophilus having appropriate acidifying and texturizing properties for dairy fermentations | |
| Jeong et al. | Characterization and antibacterial activity of a novel exopolysaccharide produced by Lactobacillus kefiranofaciens DN1 isolated from kefir | |
| EP2034848B1 (en) | Streptococcus thermophilus bacterium | |
| US20170298457A1 (en) | Lactic bacterium with modified galactokinase expression for texturizing food products by overexpression of exopolysaccharide | |
| JP5898220B2 (en) | Lactic acid strains that modify texture | |
| KR20010024311A (en) | Lactobacillus helveticus bacterium having high capability of producing tripeptide, fermented milk product, and process for preparing the same | |
| JP4669835B2 (en) | Lactic acid bacteria that improve texture | |
| JP4448896B2 (en) | Method for producing fermented milk | |
| US20080274085A1 (en) | Probiotic Bifidobacterial Species | |
| JP2017006097A (en) | Method for producing fermentation product | |
| MX2012009975A (en) | Method for producing fermented food containing bifidobacterium bacteria. | |
| JP4933124B2 (en) | Lactococcus lactic acid bacteria having an immunostimulatory effect, viscous lactococcus lactic acid bacteria, and a method for producing viscous fermented milk using these in combination | |
| CN104254604B (en) | Texturizing lactic acid bacteria strain having resistance to ampicillin | |
| CN109022330B (en) | Lactococcus lactis BL19 with high proteolytic ability and capable of producing casein fragrance and application thereof | |
| CN112512325A (en) | Method for producing improved dairy products using spore-forming negative bacillus strains | |
| Erşan et al. | Use of B acillus indicus HU 36 as a probiotic culture in set‐type, recombined nonfat yoghurt production and its effects on quality | |
| CN109105491B (en) | Lactococcus lactis IMAU11823 capable of producing sticky cheese fragrance and application thereof | |
| US11272716B2 (en) | Bacteria | |
| Yasmine et al. | Characterization of dominant cultivable lactic acid bacteria isolated from west Algerian raw camel’s milk and assessment of their technological properties | |
| JP5351113B2 (en) | Method for producing fermented food containing Bifidobacterium | |
| Tuncer et al. | Properties of exopolysaccharide producer Streptococcus thermophilus ST 8. 01 isolated from homemade yoghurt | |
| ERDEM et al. | RESEARCH Use of Bacillus indicus HU36 as a probiotic culture in set-type, recombined nonfat yoghurt production and its effects on quality |
Legal Events
| Date | Code | Title | Description |
|---|---|---|---|
| WWE | Wipo information: entry into national phase |
Ref document number: 200780041774.7 Country of ref document: CN |
|
| 121 | Ep: the epo has been informed by wipo that ep was designated in this application |
Ref document number: 07820845 Country of ref document: EP Kind code of ref document: A1 |
|
| WWE | Wipo information: entry into national phase |
Ref document number: 12443903 Country of ref document: US |
|
| WWE | Wipo information: entry into national phase |
Ref document number: MX/A/2009/003651 Country of ref document: MX |
|
| NENP | Non-entry into the national phase |
Ref country code: DE |
|
| WWE | Wipo information: entry into national phase |
Ref document number: 2007820845 Country of ref document: EP |
|
| ENP | Entry into the national phase |
Ref document number: 2009116438 Country of ref document: RU Kind code of ref document: A |
|
| ENP | Entry into the national phase |
Ref document number: PI0717777 Country of ref document: BR Kind code of ref document: A2 Effective date: 20090403 |