Pulmonary nodules computer-aided diagnosis demo system.
Basically an application integrate NoduleNet & texture classification model.
⚠️ Warning: Performance of the system is still incomplete and far away from practical use.
This is a pulmonary nodules (lung nodules) computer-aided diagnosis (CADx) demo application, which cound directly raise a diagnostic suggestion based on Lung-RADS automatically from computed tomography (CT) scan.
The application has three different view:
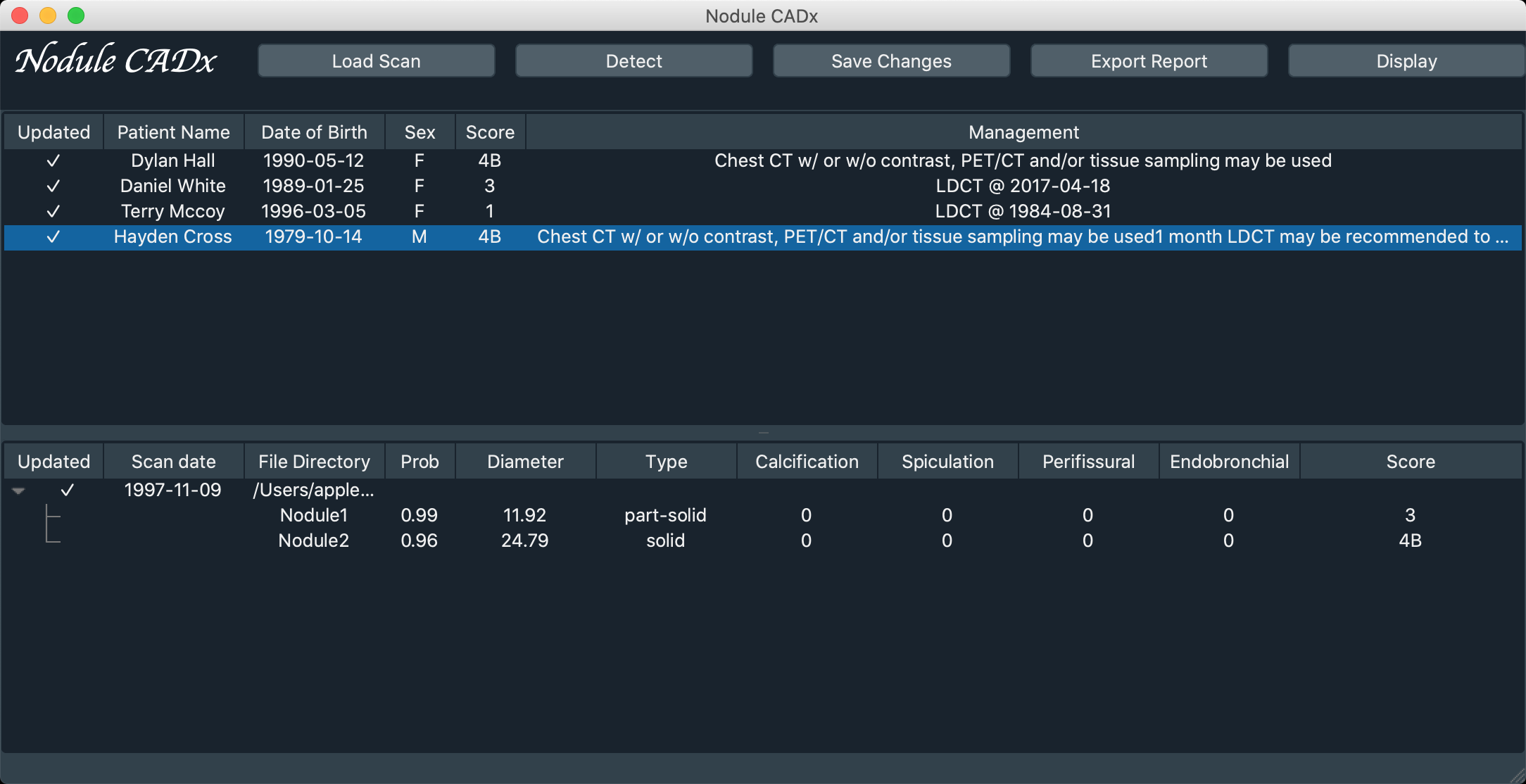
- Main Window
- Screening Window
- Preferences
- Have a quick view of patient list and corresponding scans & nodules information.
- Field introduction
- Upper block: Patient list
Updated: Indicate if all the scans of this patient have been detectedPatient Name/Date of Birth/Sex: Information of patientScore/Management: Output based on Lung-RADS
- Lower block: Scan & Nodule list
Updated: Indicate if the scan has been detectedScan date/File Directory: Information of scanType: Texture type of corresponding nodule, has five categories (Non-Solid, Non/Part, Part-Solid, Part/Solid, Solid)Calcification/Spiculation/Perifissural/Endobronchial: 0 indicate No and 1 indicate YesScore: Score of single nodule based on Lung-RADS
- Upper block: Patient list
- Button introduction
Load Scan: Load CT scanDetect: Run automatic diagnosis on seleteced scan, it may take minutesSave Changes: Since the information of nodules could be edited by user, save those changes to project file (.json)Export Report: Export field information in csv formatDisplay: Open screening window for selected scan
- Showing selected scan and it's corresponding detected nodules
- Since model isn't perfect, user can edit nodules information here
- Several simple settings of application
- LIDC-IDRI dataset label texture in 5 categories (1~5), with 2 & 4 is undetermined. Check the box will classify 2 with 1, and 4 with 5
- Threshold indicate thresholding the probability at detection stage
There are two models in models
- nodulenet_model.ckpt
- Perform nodule detection & segmentation
- Follow the training procedure describe at NoduleNet repo
- Since I modified some
data_parallelcode in net to make model run on CPU, replace the net directory for your own use may be needed
- classification_model.pkl
- Perform nodule texture classification (only for Non-Solid, Part-Solid and Solid classification)
- Train with LIDC-IDRI dataset (LUNA16 1186 nodules selected version)
- Using
Radiomicsmethodology, withPrincipal Component Analysis (PCA)andSupport Vector Regression (SVR)
Modules besides python built-ins
- GUI - PyQt5 / QDarkStyle / PyQtGraph
- Basic - NumPy / SimpleITK / OpenCV / Scikit-Image / SciPy
- Model Relatives - PyTorch / Scikit-Learn / PyRadiomics
- Others - Faker / dateutil
- Ensure you could run a NoduleNet model on your device
- Replace net directory or config.py or nodulenet_model.ckpt may be needed
- Clone the repo
git clone https://github.com/wenyalintw/Nodule_CADx.git- TODO Install required modules listed in requirements.txt
pip install -r /path/to/requirements.txt- Ready to go
cd src
python main.py- Lung ROI segmentation
- Size based on Lung-RADS criteria
- Types classification beside texture
- NoduleNet repo
- Reference of screening window apperance design: ITK-SNAP
- trashcan.png licensed under "CC BY-ND 3.0" downloaded from ICONSDB