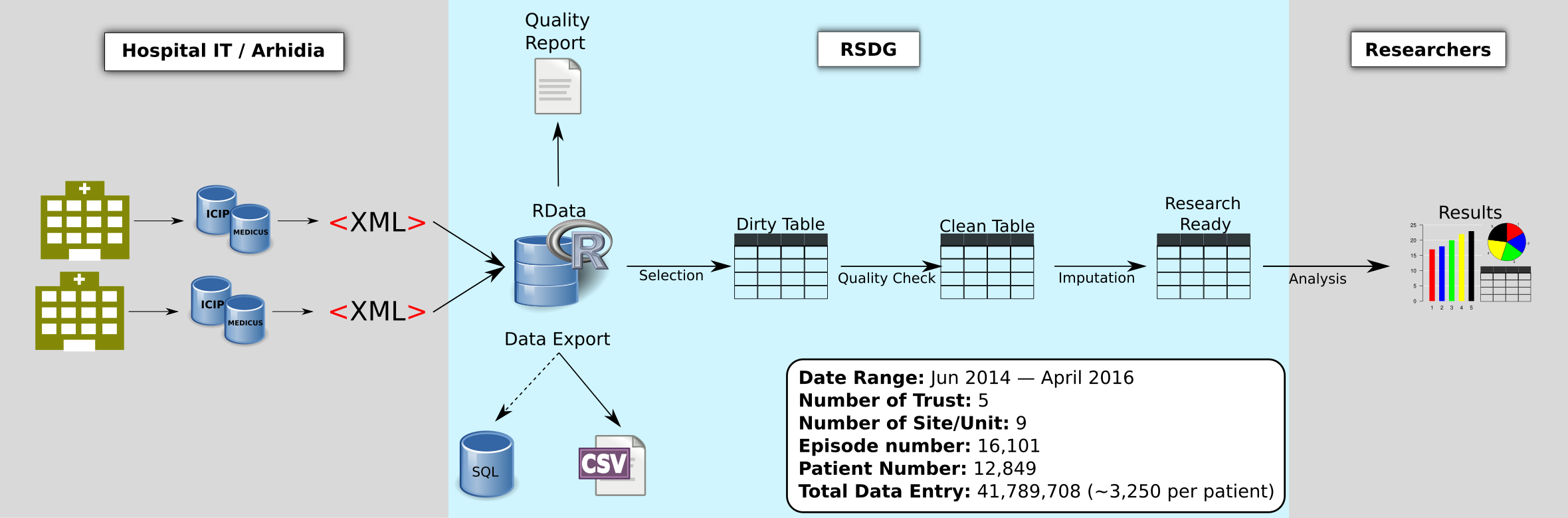
The ccdata R package is the centralised tool set for the critical care data
analysis. Three key components can be found in the current version of ccdata
package,
- The XML parser
- Data cleaning and validation modules
- Table exporter
- Data analysis functions
The ccdata package is portable to all platforms where a R environment is available.
It covers the most part of the data processing pipeline. The XML files will be
parsed to an R data structure which is a bespoken query-able storage format for
the critical patient records. With the selecting and cleaning process, the user
can obtain a clean table guided by the YAML configuration file specified by the
users. The user can subsequently perform their data analysis on the clean
table.
The YAML configuration example:
NIHR_HIC_ICU_0108:
shortName: hrate
dataItem: Heart rate
distribution: normal
decimal_places: 0
# filter1: do not use the episode where hrate cannot be found.
nodata:
apply: drop_episode
# filter2: mark all the values based on reference range (traffic colour)
# remove entries where the range check is not fullfilled.
range:
labels:
red: (0, 300)
amber: (0, 170)
green: (50, 150)
apply: drop_entry
# filter3: compute the item missing rate on given cadences; in this case, we compute the daily (red) and hourly (amber) missing rate, and only accpet episodes of which hourly missing rate (amber) is lower than 30%.
missingness:
labels:
red: 24
amber: 1
accept_2d:
amber: 70
apply: drop_episode - R (>= 2.1.0),
- XML,
- reshape2,
- data.table,
- yaml,
- pander,
- RPostgreSQL,
- sqldf
git clone [email protected]:UCL-HIC/ccdata.git
R CMD INSTALL ccdata # "sudo R CMD INSTALL ccdata" if root access is required.
- Download the
tarfile from ccdata Github page. - In the
packagepanel click the buttoninstall. - select
Package Archive File (.tgz, .tar.gz)for theselect fromtab. - click install
The ccdata package is currently underdevelopment. We wellcome users using,
commenting about the code on the master branch. If you have any question, you
can just raise an isssue on Github or contact the developers via email
([email protected]). Please let us know if you also want make contribution to the
code development.