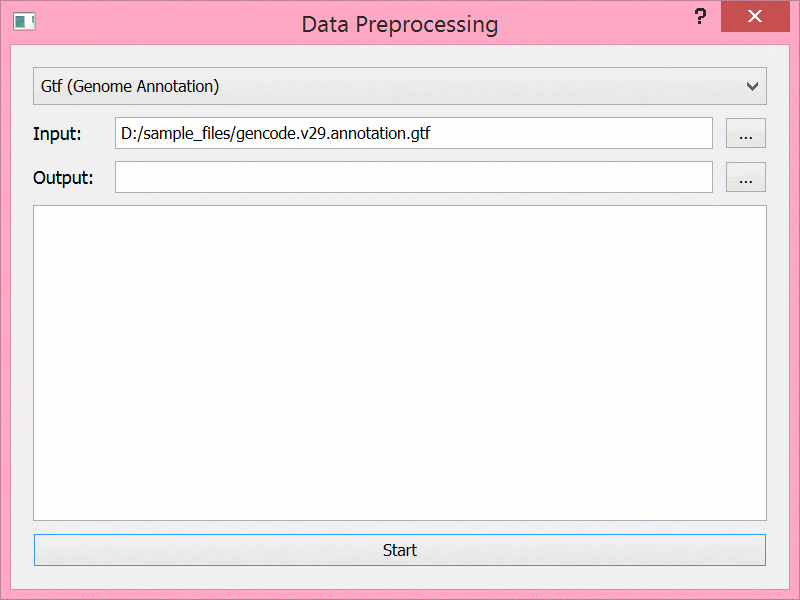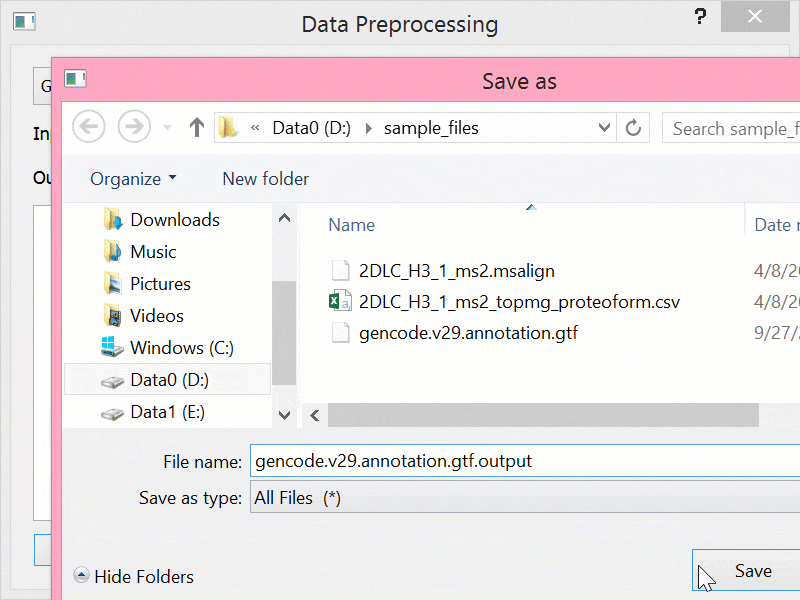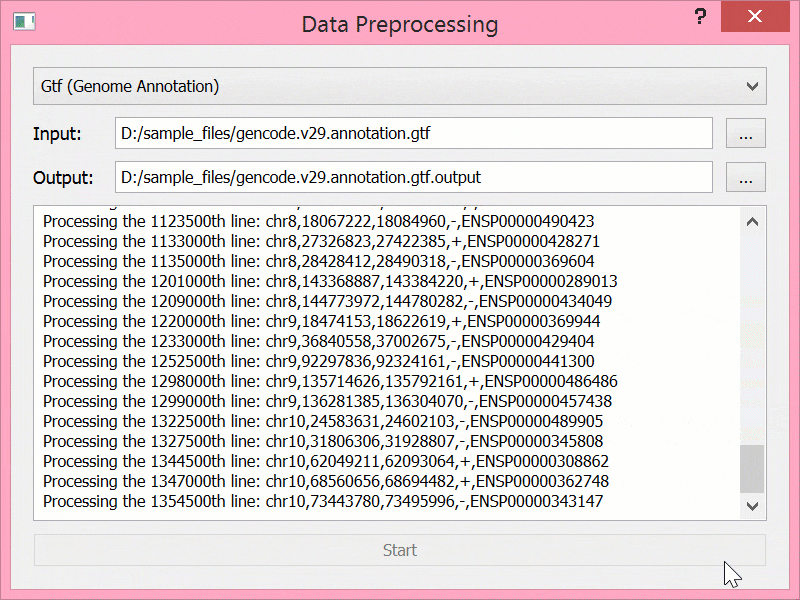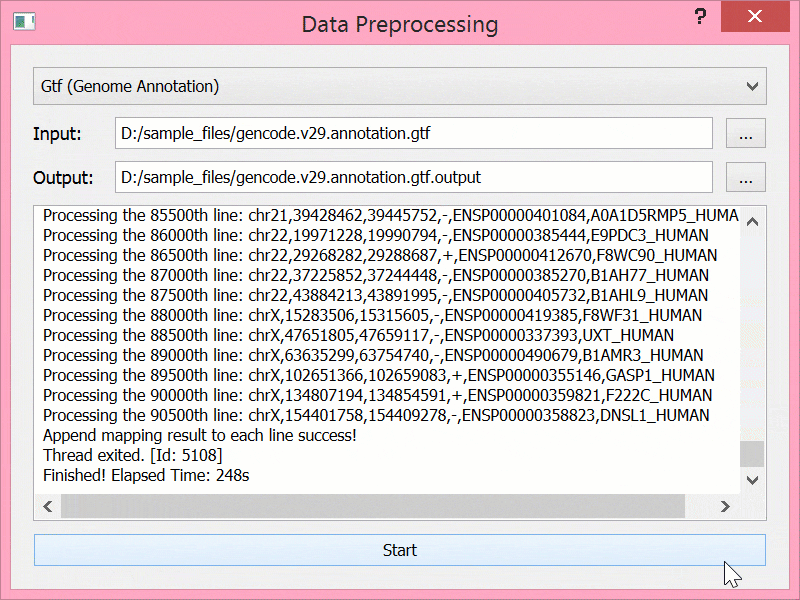
BEYONDGBrowse是一个基于jBrowse基因组浏览器的多组学序列数据可视化软件:
- 整合蛋白质组学数据,实现基因组、转录组、蛋白质组数据在同一浏览器中可视化
- 提供后端服务,支持数据进行预处理
- 支持蛋白质氨基酸序列、蛋白质质谱多样本差异分析
- 支持蛋白质组数据全基因组快速定位
- 支持蛋白质变体的翻译后修饰可视化
- 实现了高效的蛋白质变体序列、质谱数据集线性模糊匹配算法
- 蛋白质翻译、变体序列支持在线快捷注释
- 注释支持多用户、版本管理与快速检索
- 使用Linux、MacOS或装有WSL的Windows操作系统
- 如需修改Web服务器文件,例如/var/www,则需要Sudo权限
- 有一定命令行操作经验
-
Ubuntu/WSL/Debian系统:
sudo apt install git build-essential zlib1g-dev -
CentOS/RedHat:
sudo yum groupinstall "Development Tools" sudo yum install git zlib-devel perl-ExtUtils-MakeMaker
git clone https://github.com/gmod/jbrowse jbrowse
cd jbrowse
git checkout 1.16.8-release # 指定v1.16.8版本
./setup.sh
对于中国大陆用户,推荐在执行上述命令之前,使用npm镜像
npm config set registry http:https://r.cnpmjs.org
npm config set puppeteer_download_host=http:https://cnpmjs.org/mirrors
export ELECTRON_MIRROR="http:https://cnpmjs.org/mirrors/electron/"
-
切换到jbrowse根目录
cd jbrowse mkdir data && cd data -
下载参考基因组序列Fasta文件(GRCh38.p12.genome.fa)
-
生成fai索引:
samtools faidx GRCh38.p12.genome.fa -
下载样本Bam文件(GRCh38.illumina.blood.1.bam)
-
生成Bai索引:
samtools index GRCh38.illumina.blood.1.bam -
创建配置文件,
touch tracks.conf,写入保存:[GENERAL] refSeqs = GRCh38.p12.genome.fa.fai refSeqSelectorMaxSize = 1000 [tracks.refseq] urlTemplate = GRCh38.p12.genome.fa storeClass = JBrowse/Store/SeqFeature/IndexedFasta type = JBrowse/View/Track/Sequence key = _reference_sequence label = _reference_sequence showTranslation = false [tracks.alignments] urlTemplate = GRCh38.illumina.blood.1.bam storeClass = JBrowse/Store/SeqFeature/BAM type = JBrowse/View/Track/Alignments2 key = sample_alignments label = sample_alignments -
jbrowse/data目录结构:
jbrowse/data/ ├── GRCh38.illumina.blood.1.bam ├── GRCh38.illumina.blood.1.bam.bai ├── GRCh38.p12.genome.fa ├── GRCh38.p12.genome.fa.fai └── tracks.conf
-
Method1:
# 启动express.js开发服务器并监听8082端口 npm run start -
Method2:
# Nginx、Apache等其他Web服务器 sudo chown -R `whoami` <jBrowse目录> sudo mv <jBrowse目录> <Web服务器根目录,例如/var/www>
http:https://jbrowse.org/docs/installation.html
http:https://jbrowse.org/docs/tutorial.html
-
下载
# 切换到jBrowse插件目录 cd jbrowse/plugins git clone https://github.com/penguin806/BEYONDGBrowsePlugin.git BEYONDGBrowsePlugin -
激活
# 编辑jbrowse根目录配置文件jbrowse.conf,在末尾加入以下内容 [ plugins.BEYONDGBrowse ] location = plugins/BEYONDGBrowse -
配置
-
全局配置文件
jbrowse_conf.json# 编辑jbrowse根目录jbrowse_conf.json,在json对象中写入属性,例如: { "BEYONDGBrowseBackendAddr" : "localhost", //后端服务的IP地址 "BEYONDGBrowseUsername" : "Snow", // 在线注释使用的用户名 "massSpectraTrackNum" : 5 //默认质谱轨道数目 } -
轨道配置文件
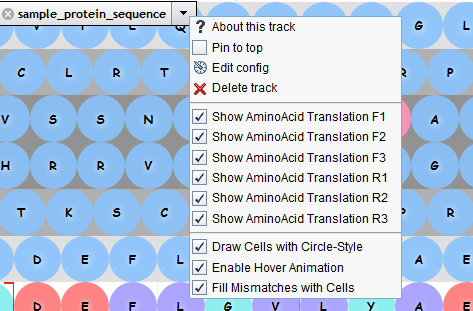
tracks.conf# 编辑data/tracks.conf,在末尾加入配置: # 参数urlTemplate = 分别对应实际数据文件GRCh38.p12.genome.fa、GRCh38.illumina.blood.1.bam [tracks.BEYONDGBrowseProteinTrack] urlTemplate = GRCh38.p12.genome.fa storeClass = JBrowse/Store/SeqFeature/IndexedFasta type = BEYONDGBrowse/View/Track/SnowSequenceTrack key = sample_protein_sequence label = sample_protein_sequence BEYONDGBrowseProteinTrack = true drawCircle = true animationEnabled = true showTranslation1st = true showTranslation2nd = true showTranslation3rd = true showTranslationReverse1st = true showTranslationReverse2nd = true showTranslationReverse3rd = true [tracks.alignments_protein] storeClass = JBrowse/Store/SeqFeature/BAM urlTemplate = GRCh38.illumina.blood.1.bam type = JBrowse/View/Track/Alignments2 glyph = BEYONDGBrowse/View/FeatureGlyph/AlignmentProteinGlyph key = sample_alignments_protein label = sample_alignments_protein
-
-
重新编译并启动
# 切换到jBrowse根目录 npm run build npm run start
- MySQL >= v5.7
-
Ubuntu:
sudo apt install -y mysql-server mysql-client sudo systemctl start mysql-server -
CentOS 7:
wget https://dev.mysql.com/get/mysql80-community-release-el7-3.noarch.rpm sudo rpm -ivh mysql80-community-release-el7-3.noarch.rpm sudo yum install -y mysql-server mysql-client sudo systemctl start mysqld -
CentOS 8:
wget https://dev.mysql.com/get/mysql80-community-release-el8-1.noarch.rpm sudo rpm -ivh mysql80-community-release-el8-1.noarch.rpm sudo yum install -y mysql-server mysql-client sudo systemctl start mysqld -
Windows: Download Community Version
-
-
下载已编译程序
访问Github Release Page,下载对应系统架构的软件包,解压缩
-
源码编译
# OS: Ubuntu / WSL on Windows sudo apt update sudo apt install -y build-essential git sudo apt install -y qt5-default qtbase5-private-dev # Since QtHttpServer is a Beta feature in Qt-Lab until now, manually compile and install the module is needed git clone --recursive https://github.com/qt-labs/qthttpserver.git qthttpserver cd qthttpserver qmake && make && make install cd .. mkdir build git clone https://github.com/penguin806/BEYONDGBrowsePreprocessor.git BEYONDGBrowsePreprocessor cd BEYONDGBrowsePreprocessor qmake && make && cp GtfFilePreprocessor ../build cd .. git clone https://github.com/penguin806/BEYONDGBrowseDatabaseImport.git BEYONDGBrowseDatabaseImport cd BEYONDGBrowseDatabaseImport qmake && make && cp BEYONDGBrowseDatabaseTool ../build cd .. git clone https://github.com/penguin806/BEYONDGBrowseBackend.git BEYONDGBrowseBackend cd BEYONDGBrowseBackend qmake && make && cp SnowPluginBackend config.ini ../build cd ..build # If all build is successful, the file structure looks like the following # build/ # ├── BEYONDGBrowseDatabaseImport # ├── config.ini # ├── GtfFilePreprocessor # └── SnowPluginBackend
-
编辑config.ini
[General] ; 后端监听端口 listenPort=12080 [Database] ; MySQL数据库主机地址 serverAddrress=localhost ; MySQL数据库名 databaseName=beyondgbrowse ; MySQL用户名 username=snow_db201905 ; MySQL用户密码 password=snow_db201905
sample_files/
├── 2DLC_H3_1_ms2.msalign
├── 2DLC_H3_1_ms2_topmg_proteoform.csv
└── gencode.v29.annotation.gtf
-
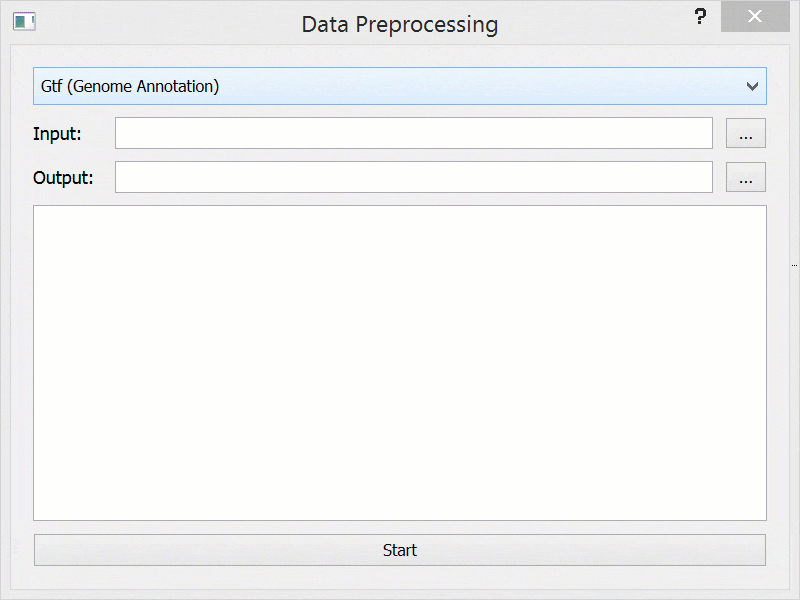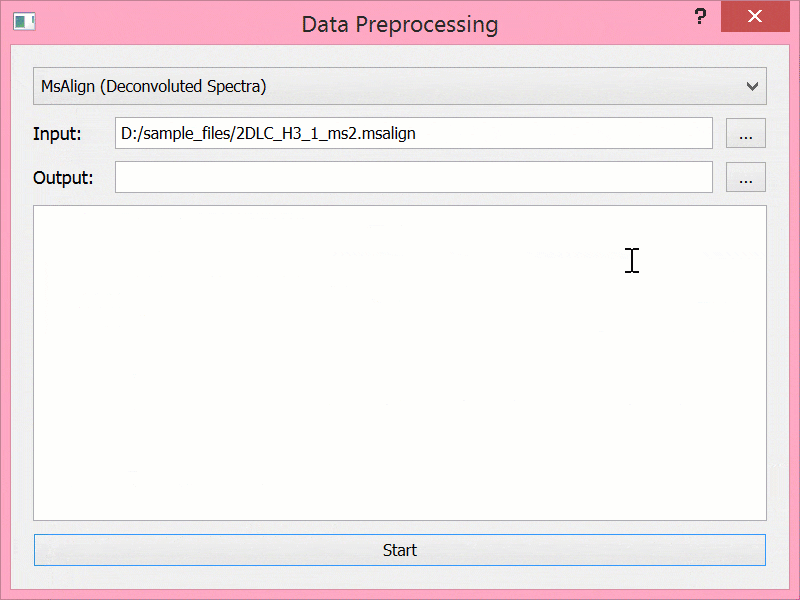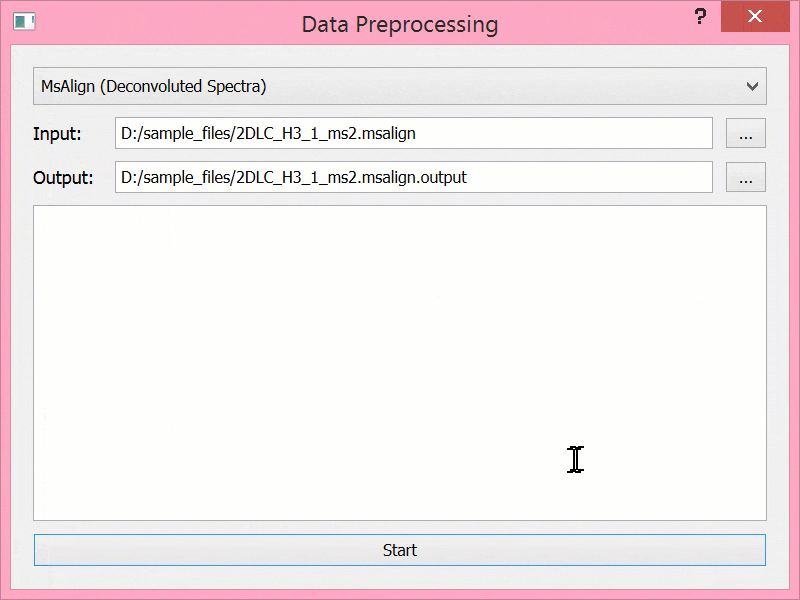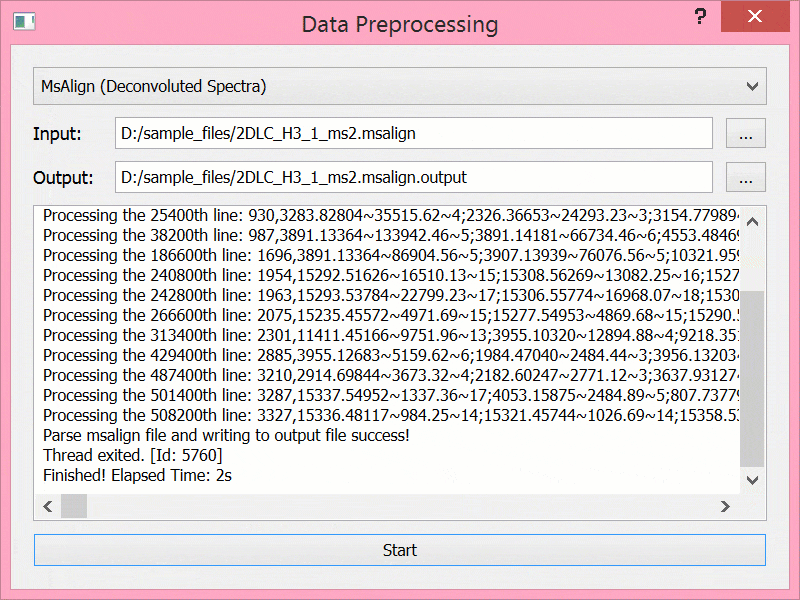
预处理结束后文件结构
sample_files/ ├── 2DLC_H3_1_ms2.msalign ├── 2DLC_H3_1_ms2.msalign.output ├── 2DLC_H3_1_ms2_topmg_proteoform.csv ├── gencode.v29.annotation.gtf ├── gencode.v29.annotation.gtf.output └── gencode.v29.annotation.gtf.output.tmp
浏览器访问http:https://<jBrowse URL>/
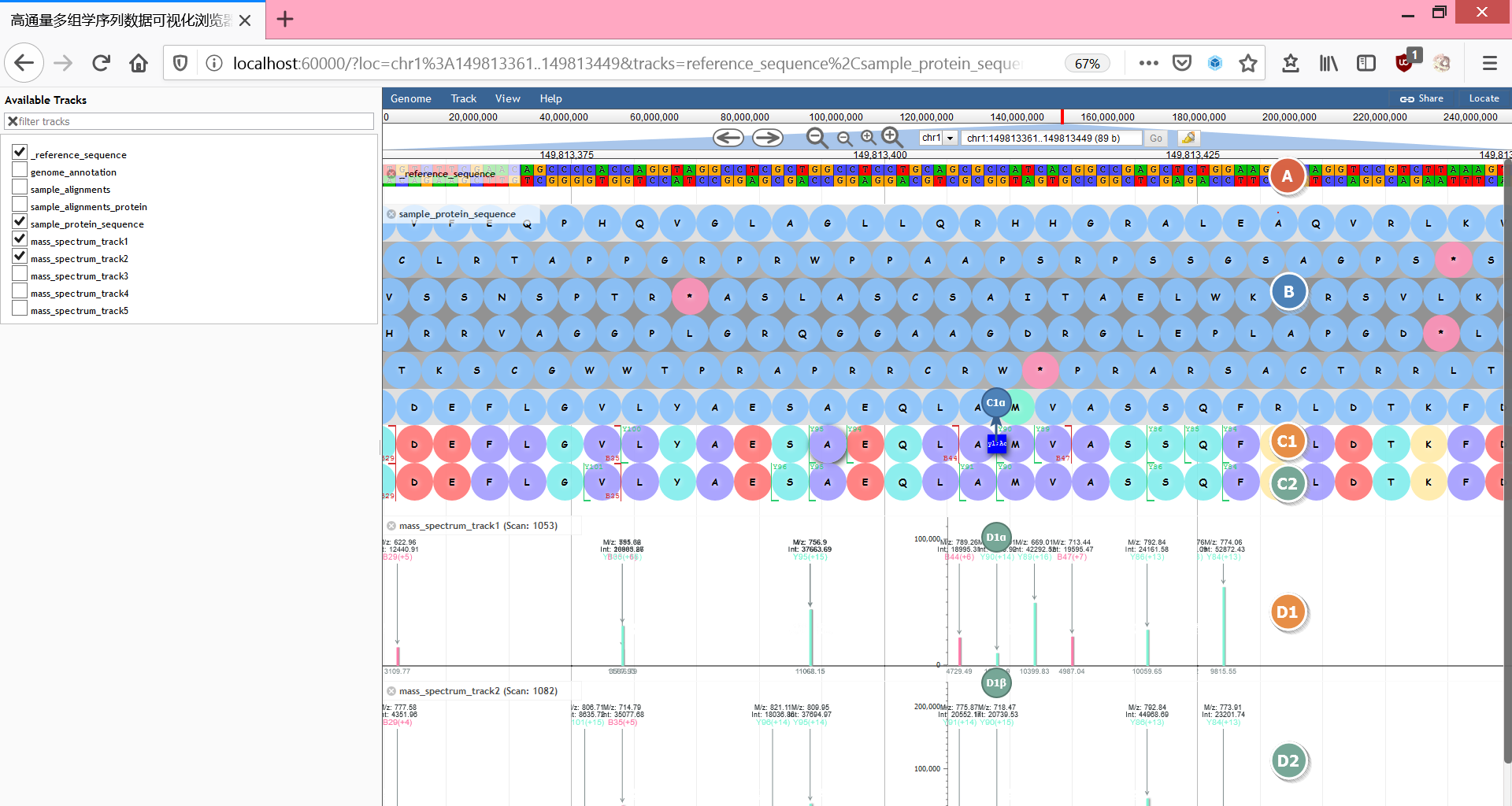
- A. 参考基因序列
- B. 参考蛋白质翻译
- C1. 样本1蛋白质变体序列
- C1α. 蛋白质变体修饰
- C2. 样本2蛋白质变体序列
- D1. 样本1质谱
- D1α. 峰度
- D1β. 质量
- D2. 样本2质谱
质谱红色/绿色,分别代表B离子/Y离子
蛋白质变体颜色含义:
- 绿色 (
#81ecece6): 亲水性- 紫色 (
#a29bfee6): 疏水性- 红色 (
#ff7675e6): 酸性- 黄色 (
#ffeaa7e6): 碱性
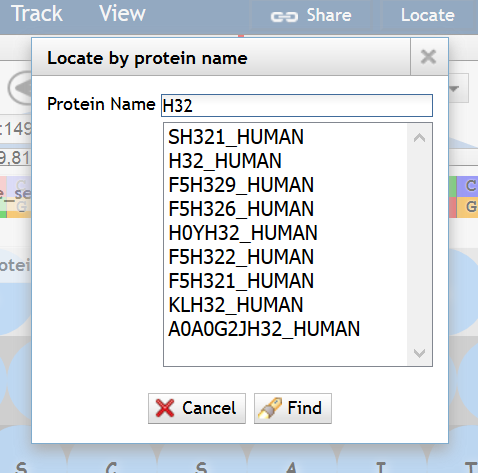
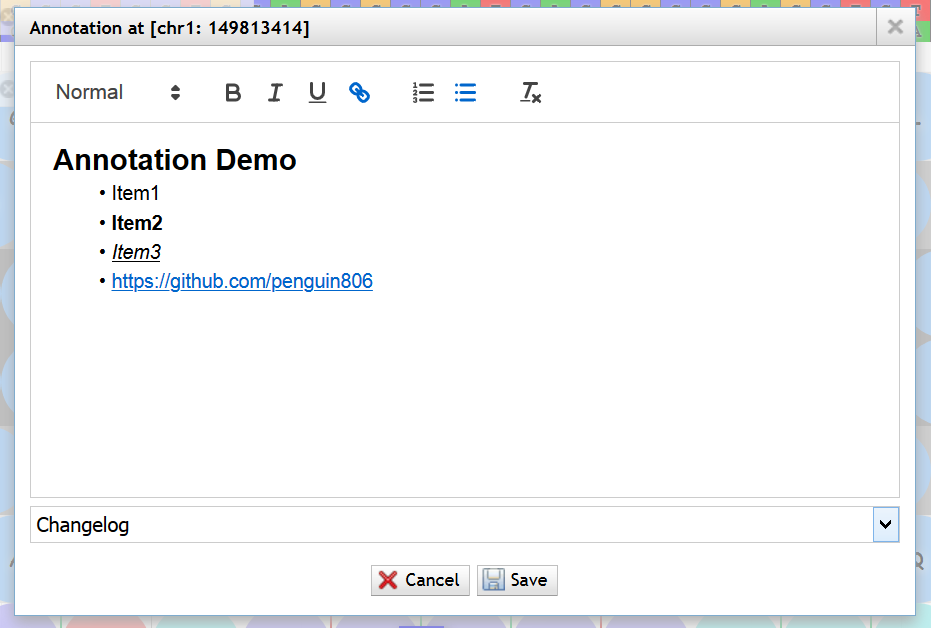
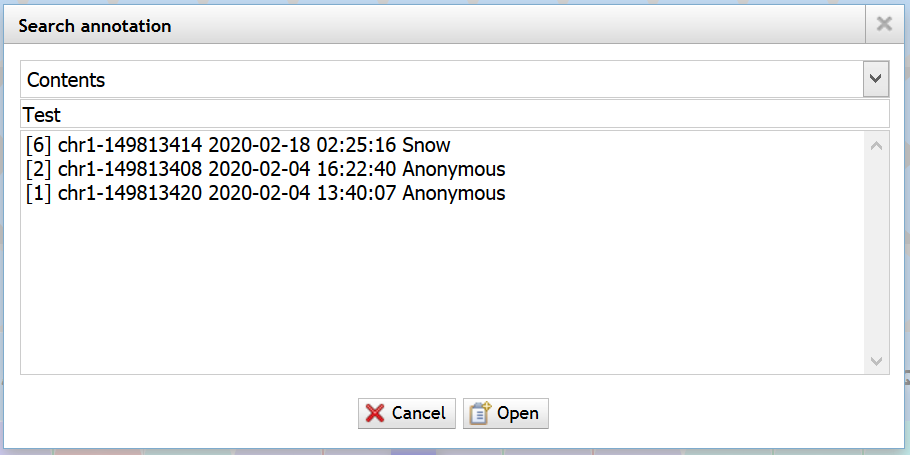
顶部菜单<Track> -> <Search annotation>
支持以 内容、ID、作者 或 IP地址 作为条件查找
- 钟坚成教授 湖南师范大学 邮箱:[email protected]
- 学生:
- 李学锋 湖南师范大学 邮箱:[email protected]