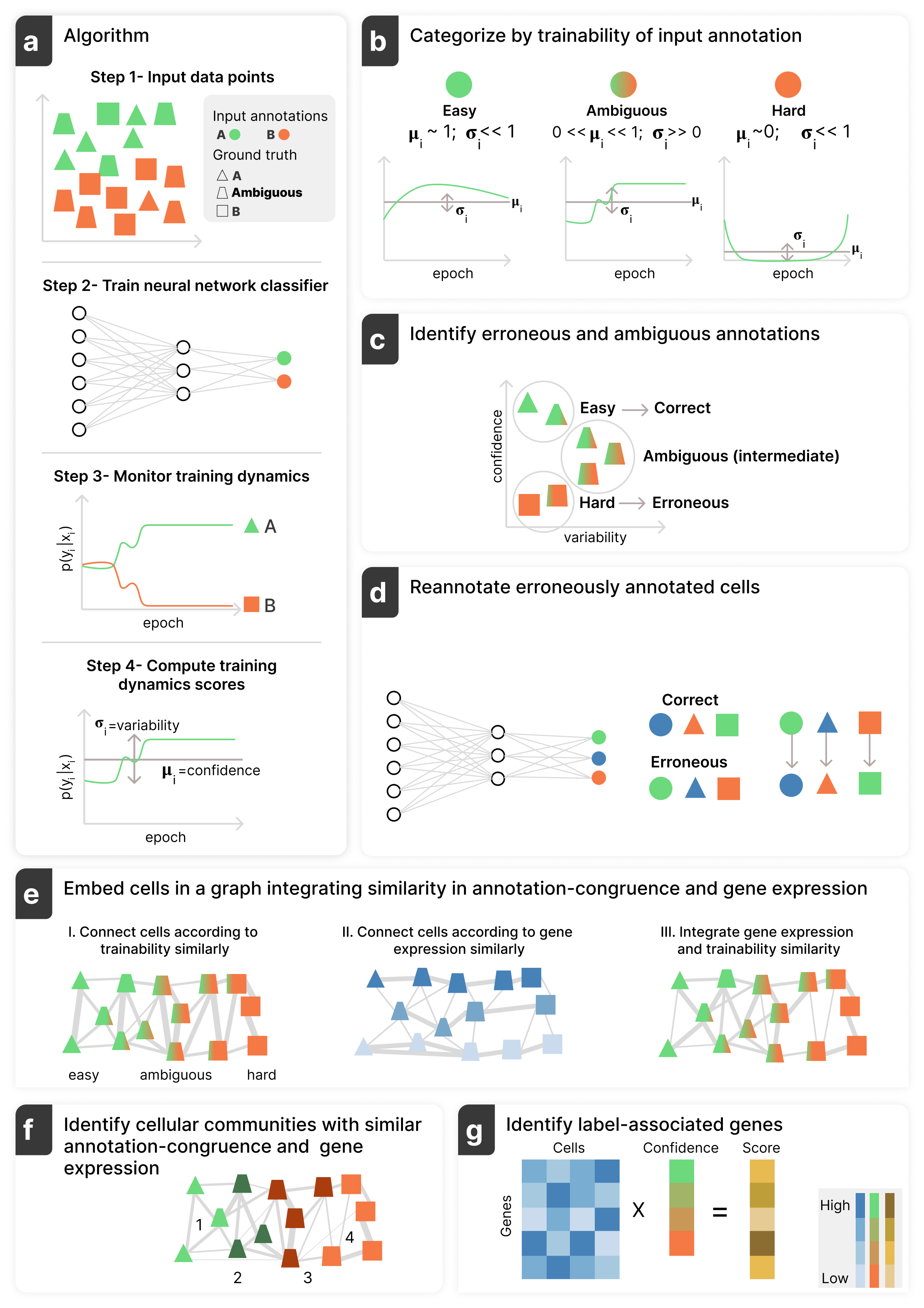
Annotatability, a method to identify meaningful patterns in single-cell genomics data through annotation-trainability analysis, which estimates annotation congruence using a rich but often overlooked signal, namely the training dynamics of a deep neural network.
For reproducibility of Annotability manuscript, please refer to:
https://github.com/nitzanlab/Annotatability_notebooks
git clone https://github.com/nitzanlab/Annotatability.git
cd Annotatability
pip install .Install time- depends on the installation time of Pytorch, a few minutes on a normal computer.
The code is based on Scanpy package.
An example of the usage of our method is available in the following [tutorial] (https://github.com/nitzanlab/Annotatability_notebooks/blob/main/tutorial_retina.ipynb
) (runtime of a few minutes with GPU, ~20 minutes without GPU).
Annotatability comprises two code files:
"models.py", which encompasses the training of neural network functions and the generation of the trainability-aware graph.
"metrics.py", which contains the scoring functions.
Imports:
from Annotatability import metrics, models
import numpy as np
import scanpy as sc
from torch.utils.data import TensorDataset, DataLoader , WeightedRandomSampler
import torch
import torch.optim as optim
device = torch.device('cuda:0' if torch.cuda.is_available() else 'cpu')
Train the neural network and calculate the confidence and variability metrics
We take as input annotated data (of type Anndata) named “adata”, and the annotation “label” (stores as observation) we aim to analyze.
To estimate the confidence and variability of the annotation of each cell, we use the following commands:
epoch_num=50 %Can be changed
prob_list = models.follow_training_dyn_neural_net(adata, label_key='label',iterNum=epoch_num)
all_conf , all_var = models.probability_list_to_confidence_and_var(prob_list, n_obs= adata.n_obs, epoch_num=epoch_num, device=device)
adata.obs["var"] = all_var.detach().numpy()
adata.obs["conf"] = all_conf.detach().numpy()
For 'follow_training_dyn_neural_net' function, we can change the following hyperparameters-
iterNum : int, optional (default=100)
Number of training iterations (epochs).
lr : float, optional (default=0.001)
Learning rate for the optimizer.
momentum : float, optional (default=0.9)
Momentum for the optimizer.
device : str, optional (default='cpu')
Device for training the neural network ('cpu' or 'cuda' for GPU).
weighted_sampler : bool, optional (default=True)
Whether to use a weighted sampler for class imbalance.
batch_size : int, optional (default=256)
Batch size for training.
num_layers : int, optional (default=3)
Depth of the neural network. Values alowed=3/4/5
Compute the annotation-trainability score
adata_ranked = metrics.rank_genes_conf_min_counts(adata)
The results will be stored as variables in:
adata_ranked.var['conf_score_high'] %annotation-trainability positive association score
adata_ranked.var['conf_score_low'] %annotation-trainability negative association score
Trainability-aware graph embedding
connectivities_graph , distance_graph = metrics.make_conf_graph(adata.copy(), alpha=0.9 , k=15)
adata.obsp['connectivities']=sp.csr_matrix(connectivities_graph)
Note: 'alpha' can be adjusted.
For visualization of the trainability-aware graph you can use the following functions:
sc.tl.umap(adata)
sc.pl.umap(adata, color='conf')
Notice that using sc.pp.neighbors(adata) will store the neighbors graph in adata.obsp['connectivities'] instead of the trainability-aware graph.
python (>3.0)
packages:
"numpy",
"scanpy",
"numba",
"pandas",
"scipy",
"matplotlib",
"pytest",
"torch"
conda create -n Annotatability python=3.10
conda activate Annotatability
pip install .
pytest tests/test
Jonathan Karin - jonathan.karin [at ] mail.huji.ac.il