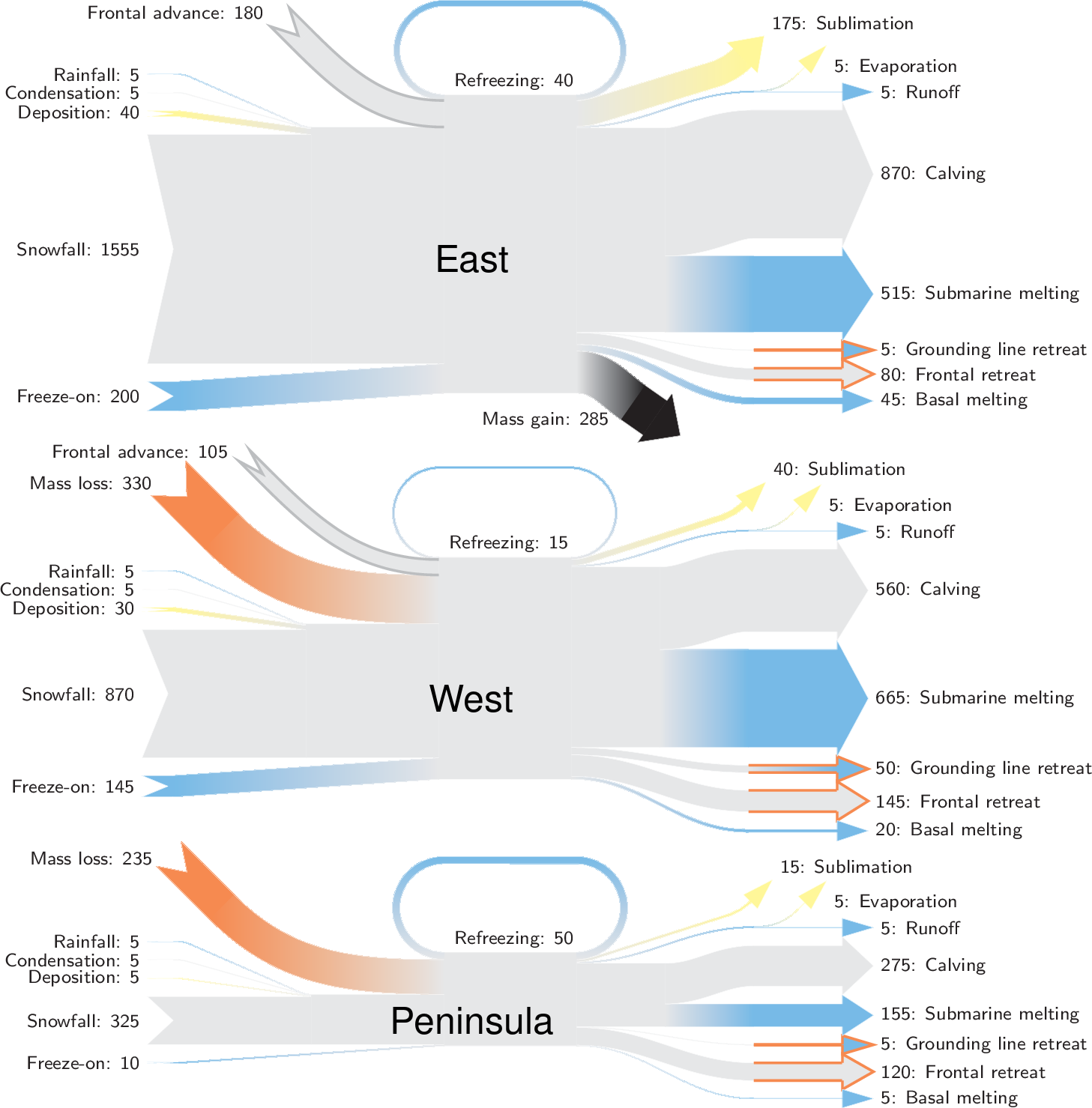
Sankey diagrams for mass flow in Greenland and Antarctica.
See ./ms.tex
Note
All figures are width-proportional within and between each other.
| Code | Term | Value | I/O | Period | Source | Comment |
|---|---|---|---|---|---|---|
| RF | Rainfall | 45 | I | 2000-2019 | fettweis_2020 | |
| CD | Condensation | 5 | I | 2000-2019 | fettweis_2020 | |
| DP | Deposition | 10 | I | 2000-2019 | fettweis_2020 | |
| SF | Snowfall | 685 | I | 2000-2019 | fettweis_2020 | |
| RFZ | Refreezing | 195 | IO | 2000-2019 | fettweis_2020 | Melt + rainfall - runoff |
| EV | Evaporation | 10 | O | 2000-2019 | fettweis_2020 | |
| RU | Runoff | 440 | O | 2000-2019 | fettweis_2020 | |
| BM | Basal melting | 20 | O | steady | karlsson_2021 | |
| DYN | Discharge | 475-25 | - | 2000-2019 | mankoff_2021_solid,kochtitzky_2023 | Submarine melting + calving - SMB |
| SUB | Submarine melting | 225 | O | enderlin_2013 | 50 % of discharge | |
| SUBF | Freeze-on | 0 | I | None in Greenland | ||
| ICE | Calving | 225 | O | enderlin_2013 | 50 % of discharge | |
| GZRET | Grounding line retreat | 5 | O | Estimate | ||
| FRLOSS | Frontal retreat | 50 | O | 2000-2020 | kochtitzky_2023 | |
| FRGAIN | Frontal advance | 0 | O | None in Greenland | ||
| SU | Sublimation | 60 | O | 2000-2019 | fettweis_2020 | |
| DD | Mass loss | - | Derived | sum(O) - sum(I) | ||
| MG | Mass gain | - | Derived | sum(I) - sum(O) |
trash G_GL
[[ -e ./G_GL ]] || grass -e -c EPSG:3413 ./G_GLcat ~/data/Mankoff_2021/708/MB_SMB_D_BMB_ann.csv \
| cut -d, -f1,6 \
| sed -e '1,/^1999/d' \
| sed '/^2019/q' \
| datamash -t, mean 1,22009.5,476.44290795 org_babel_sh_eoe
Then, subtract 25 from 475 based on citet:kochtitzky_2023 who report, in Section 3.3, 17 +- 6.8 and 14.5 +- 5.8 but that “[b]ecause our fluxgates were typically located tens to hundreds of meters lower than those in the similar studies (King et al., 2018; Mankoff et al., 2020), the melt correction for these studies would be higher than values presented herein, although it is beyond the scope of the current study to determine what those values would be.”
- 21 Gt/yr from Karlsson (2021) https://doi.org/10.1038/s41467-021-23739-z
- Assume steady state
From Millan (2022) https://doi.org/10.5194/tc-16-3021-2022
- Gz retreat is ~0.13 km/yr (Fig. 3a)
- Ice velocity is ~1200 m/yr (Fig. 3b) (not needed)
- 20 km wide
Rates are higher per Ciraci (2023) https://doi.org/10.1073/pnas.2220924120, but
- Ice surface close to flotation near GZ, and shelf is ~500 m thick, so estimate 600 m ice.
Therefore, gz retreat in Gt/year is width * thick * retreat rate * density
frink "0.13 km/yr * 20 km * 600 m * 917 kg/m^3 -> Gt/yr"1.43052
Assume similar from other ice shelves too, for a total of ~5 Gt/yr GZ retreat in Greenland.
g.mapset -c MAR
ncdump -v TIME dat/MARv3.12-GRD-15km-annual.nc4 # 20-39 = 2000-2019
ncra --overwrite -d TIME,20,39 dat/MARv3.12-GRD-15km-annual.nc4 tmp/MAR_GL.nc
ncdump -v X10_110 tmp/MAR_GL.nc # 101
ncdump -v Y20_200 tmp/MAR_GL.nc # 181
g.region w=$(( -645000 - 7500 )) e=$(( 855000 + 7500 )) s=$(( -3357928 - 7500 )) n=$((-657928 + 7500 )) res=15000 -p
var=SF # debug
for var in SF RF RU SU ME SMB EVA CON DEP SUB MSK AREA; do
r.in.gdal -o input=NetCDF:tmp/MAR_GL.nc:${var} output=${var}
r.region -c map=${var}
done
r.mapcalc "GL_ice_all = (MSK > 50) & ((x()-y()) > 520000)" # Limit to ice and remove Canada
r.clump input=GL_ice output=clumps --o
main_clump=$(r.stats -c -n clumps sort=desc | head -n2 | tail -n1 | cut -d" " -f1)
r.mapcalc "GL_ice = if(clumps == ${main_clump}, 1, null())"
r.mask raster=GL_ice --o
# scale
## units are mm.w.eq. per grid cell. Grid cell areas are in km^2
## + mm.w.eq. -> m w.eq.: /1E3
## + m w.eq -> kg: *1E3
## + area in km^2 -> m^2: *1E3*1E3
## + kg -> Gt: /1E12
# ds = ds/1E3 * 1E3 * ds['AREA']*1E3*1E3 / 1E12
for var in SF RF RU SU ME SMB EVA CON DEP SUB; do
r.mapcalc "${var} = (${var}/1000) * 1000 * (AREA * 1000*1000) / exp(10,12)"
done
r.mask -r
r.mapcalc "RFZ = ME + RF - RU"for var in SF RF RU ME SMB EVA CON DEP SUB RFZ; do
echo ${var} $(r.univar -g ${var} | grep sum)
doneSF sum=686.768815213334 RF sum=45.5535346610575 RU sum=440.665680238757 ME sum=589.542715610605 SMB sum=235.536411205988 EVA sum=7.9188290228966 CON sum=2.15906279235185 DEP sum=12.2697684982692 SUB sum=61.8983408836194 RFZ sum=194.430570032905
import pandas as pd
df = pd.read_csv('/home/kdm/data/Mankoff_2020/ice/GIS_D.csv', index_col=0, parse_dates=True)
df = df['2000-01-01':'2019-12-31']
df.resample('YS').mean().mean().round().astype(int).values[0]
487
| Code | Term | Value | I/O | Period | Source | Comment |
|---|---|---|---|---|---|---|
| RF | Rainfall | 5 | I | 2000-2019 | fettweis_2020 | |
| CD | Condensation | 5 | I | 2000-2019 | fettweis_2020 | |
| DP | Deposition | 75 | I | 2000-2019 | fettweis_2020 | |
| SF | Snowfall | 2750 | I | 2000-2019 | fettweis_2020 | |
| RFZ | Refreezing | 105 | IO | 2000-2019 | fettweis_2020 | |
| EV | Evaporation | 5 | O | 2000-2019 | fettweis_2020 | |
| RU | Runoff | 10 | O | 2000-2019 | fettweis_2020 | |
| BM | Basal melting | 70 | O | - | van-liefferinge_2013 | |
| DYN | Discharge | 1335+1350+(2275-75-1840) | - | 1997-2021; 1999-2017 | Sum of SUB + ICE | See caption |
| SUB | Submarine melting | 1335 | O | 2000-2017 | paolo_2023 | |
| SUBF | Freeze-on | 355 | I | 2000-2017 | paolo_2023 | |
| ICE | Calving | 1350+(2275-75-1840) | O | 1997-2021; 1999-2017 | davison_2023 + rignot_2019 grounded | See caption |
| GZRET | Grounding line retreat | 50 | O | 1997-2021 | Davison (personal comm.) | |
| FRLOSS | Frontal retreat | 79+122+145-1 | O | 2000-2021 | greene_2022 | |
| FRGAIN | Frontal advance | 181+1+103 | O | 2000-2021 | greene_2022 | |
| SU | Sublimation | 230 | O | 2000-2019 | fettweis_2020 | |
| DD | Drawdown | - | Derived | sum(O) - sum(I) | ||
| MG | Mass gain | - | Derived | sum(I) - sum(O) |
| Code | Term | Value | I/O | Period | Source | Comment |
|---|---|---|---|---|---|---|
| RF | Rainfall | 5 | I | 2000-2019 | fettweis_2020 | |
| CD | Condensation | 5 | I | 2000-2019 | fettweis_2020 | |
| DP | Deposition | 40 | I | 2000-2019 | fettweis_2020 | |
| SF | Snowfall | 1555 | I | 2000-2019 | fettweis_2020 | |
| RFZ | Refreezing | 40 | IO | 2000-2019 | fettweis_2020 | |
| EV | Evaporation | 5 | O | 2000-2019 | fettweis_2020 | |
| RU | Runoff | 5 | O | 2000-2019 | fettweis_2020 | |
| BM | Basal melting | 45 | O | - | van-liefferinge_2013 | |
| DYN | Discharge | 515+680+(1100-910) | - | 1997-2021; 1999-2017 | Sum of SUB + ICE | See caption |
| SUB | Submarine melting | 515 | O | 2000-2017 | paolo_2023 | |
| SUBF | Freeze-on | 200 | I | 2000-2017 | paolo_2023 | |
| ICE | Calving | 680 + (1100-910) | O | 1997-2021; 1999-2017 | davison_2023 + rignot_2019 grounded | See caption |
| GZRET | Grounding line retreat | 5 | O | 1997-2021 | Davison (personal comm.) | |
| FRLOSS | Frontal retreat | 80 | O | 2000-2021 | greene_2022 | |
| FRGAIN | Frontal advance | 180 | O | 2000-2021 | greene_2022 | |
| SU | Sublimation | 175 | O | 2000-2019 | fettweis_2020 | |
| DD | Drawdown | - | Derived | sum(O) - sum(I) | ||
| MG | Mass gain | - | Derived | sum(I) - sum(O) |
| Code | Term | Value | I/O | Period | Source | Comment |
|---|---|---|---|---|---|---|
| RF | Rainfall | 5 | I | 2000-2019 | fettweis_2020 | |
| CD | Condensation | 5 | I | 2000-2019 | fettweis_2020 | |
| DP | Deposition | 30 | I | 2000-2019 | fettweis_2020 | |
| SF | Snowfall | 870 | I | 2000-2019 | fettweis_2020 | |
| RFZ | Refreezing | 15 | IO | 2000-2019 | fettweis_2020 | |
| EV | Evaporation | 5 | O | 2000-2019 | fettweis_2020 | |
| RU | Runoff | 5 | O | 2000-2019 | fettweis_2020 | |
| BM | Basal melting | 20 | O | - | van-liefferinge_2013 | |
| DYN | Discharge | 665 + 560 + (765-765) | - | 1999-2017 | Sum of SUB + ICE | See caption |
| SUB | Submarine melting | 665 | O | 2000-2017 | paolo_2023 | |
| SUBF | Freeze-on | 145 | I | 2000-2017 | paolo_2023 | |
| ICE | Calving | 560 + (765-765) | O | 1997-2021; 1999-2017 | davison_2023 + rignot_2019 grounded | See caption |
| GZRET | Grounding line retreat | 50 | O | 1997-2021 | Davison (personal comm.) | |
| FRLOSS | Frontal retreat | 145 | O | 2000-2021 | greene_2022 | |
| FRGAIN | Frontal advance | 105 | O | 2000-2021 | greene_2022 | |
| SU | Sublimation | 40 | O | 2000-2019 | fettweis_2020 | |
| DD | Drawdown | - | Derived | sum(O) - sum(I) | ||
| MG | Mass gain | - | Derived | sum(I) - sum(O) |
| Code | Term | Value | I/O | Period | Source | Comment |
|---|---|---|---|---|---|---|
| RF | Rainfall | 5 | I | 2000-2019 | fettweis_2020 | |
| CD | Condensation | 5 | I | 2000-2019 | fettweis_2020 | |
| DP | Deposition | 5 | I | 2000-2019 | fettweis_2020 | |
| SF | Snowfall | 325 | I | 2000-2019 | fettweis_2020 | |
| RFZ | Refreezing | 50 | IO | 2000-2019 | fettweis_2020 | |
| EV | Evaporation | 5 | O | 2000-2019 | fettweis_2020 | |
| RU | Runoff | 5 | O | 2000-2019 | fettweis_2020 | |
| BM | Basal melting | 5 | O | - | van-liefferinge_2013 | |
| DYN | Discharge | 155 + 105 + (330-160) | - | 1997-2021; 1999-2017 | Sum of SUB + ICE | See caption |
| SUB | Submarine melting | 155 | O | 2000-2017 | paolo_2023 | |
| SUBF | Freeze-on | 10 | I | 2000-2017 | paolo_2023 | |
| ICE | Calving | 105 + (330 - 160) | O | 1997-2021; 1999-2017 | davison_2023 + rignot_2019 grounded | See caption |
| GZRET | Grounding line retreat | 5 | O | 1997-2021 | Davison (personal comm.) | |
| FRLOSS | Frontal retreat | 120 | O | 2000-2021 | greene_2022 | |
| FRGAIN | Frontal advance | 0 | O | 2000-2021 | greene_2022 | |
| SU | Sublimation | 15 | O | 2000-2019 | fettweis_2020 | |
| DD | Drawdown | - | Derived | sum(O) - sum(I) | ||
| MG | Mass gain | - | Derived | sum(I) - sum(O) |
Exported aq_baseline to ./dat/aq_baseline.csv
Exported aq_east to ./dat/aq_east.csv
Exported aq_west to ./dat/aq_west.csv
Exported aq_peninsula to ./dat/aq_peninsula.csv
trash G_AQ
[[ -e ./G_AQ ]] || grass -e -c EPSG:3031 ./G_AQgrass ./G_AQ/PERMANENT
v.in.ogr input=${DATADIR}/NSIDC/NSIDC-0709.002/1992.02.07/IceBoundaries_Antarctica_v02.shp output=basins
g.region vector=basins res=10000 -pas
v.db.select map=basins|head
v.db.select -c map=basins columns=Regions | sort | uniq # East West Peninsula Islands
v.db.select -c map=basins columns=TYPE | sort | uniq # FL GR IS (float, ground, island)
v.to.rast input=basins output=east use=val val=1 where='(Regions == "East")'
v.to.rast input=basins output=west use=val val=2 where='(Regions == "West")'
v.to.rast input=basins output=peninsula use=val val=3 where='(Regions == "Peninsula")'
r.patch input=east,west,peninsula output=basins
r.category basins separator=":" rules=- << EOF
1:East
2:West
3:Peninsula
EOF
r.colors map=basins color=viridisg.mapset -c MAR
ncdump -v TIME dat/MARv3.12-ANT-35km-annual.nc4 # 20-39 = 2000-2019
ncra --overwrite -d TIME,20,39 dat/MARv3.12-ANT-35km-annual.nc4 tmp/MAR_AQ.nc
ncdump -v X tmp/MAR_AQ.nc # 176
ncdump -v Y tmp/MAR_AQ.nc # 148
g.region w=$(( -3010000 - 17500 )) e=$(( 3115000 + 17500 )) s=$(( -2555000 - 17500 )) n=$(( 2590000 + 17500 )) res=35000 -p
var=SF # debug
for var in SF RF RU ME SMB EVA CON DEP SUB MSK AREA; do
r.in.gdal -o input=NetCDF:tmp/MAR_AQ.nc:${var} output=${var}
r.region -c map=${var}
done
# scale
## units are mm.w.eq. per grid cell. Grid cell areas are in km^2
## + mm.w.eq. -> m w.eq.: /1E3
## + m w.eq -> kg: *1E3
## + area in km^2 -> m^2: *1E3*1E3
## + kg -> Gt: /1E12
# ds = ds/1E3 * 1E3 * ds['AREA']*1E3*1E3 / 1E12
for var in SF RF RU ME SMB EVA CON DEP SUB; do
r.mapcalc "${var} = (${var}/1000) * 1000 * (AREA * 1000*1000) / exp(10,12)"
done
r.mapcalc "RFZ = ME + RF - RU"r.mask --o raster=basins@PERMANENT --q maskcats="1 thru 3 10 thru 20" # drop 0 and Islands
for var in SF RF RU ME SMB EVA CON DEP SUB RFZ; do
echo -n "${var}"
r.univar -gt map=${var} zones=basins@PERMANENT | cut -d"|" -f2,13 | column -s"|" -t | sed 's/label.*//'
r.univar -g ${var} | grep sum
echo "#"; echo "#"
done
r.mask -r --qSF East 1555.92838304071 West 868.756236659932 Peninsula 327.008298435155 sum=2751.6929181358 RF East 1.37427316764175 West 0.67184557194045 Peninsula 4.4182855932415 sum=6.46440433282369 RU East 3.03921478456715 West 0.036433758652 Peninsula 6.24173336942285 sum=9.317381912642 ME East 41.8875327525325 West 13.5639532884436 Peninsula 51.8076872767586 sum=107.259173317735 SMB East 1421.34893771318 West 856.678097752916 Peninsula 314.290356315015 sum=2592.31739178111 EVA East 1.3076393190111 West 0.4376933850929 Peninsula 1.3900330901803 sum=3.1353657942843 CON East 0.00461569848685 West 0.00432677288165001 Peninsula 0.0478741559012 sum=0.0568166272697001 DEP East 42.1006070552508 West 28.4439147061151 Peninsula 6.8402185663563 sum=77.384740327722 SUB East 174.090628819002 West 40.7804740506949 Peninsula 16.1757877048917 sum=231.046890574587 RFZ East 40.2225911356072 West 14.199365101732 Peninsula 49.9842395005773 sum=104.406195737917 [Raster MASK present]
Van Liefferinge (2013) https://doi.org/10.5194/cp-9-2335-2013
Convert MAT file to XYZ for importing into GRASS
import scipy as sp
import numpy as np
import pandas as pd
mat = sp.io.loadmat('/home/kdm/data/Van_Liefferinge_2023/Melt_Mean_Std_15exp.mat')
X = mat['X'].flatten() * 1E3 # convert from km to m
Y = mat['Y'].flatten() * 1E3
m = mat['MeanMelt'].flatten() / 10 # cm to mm
e = mat['StdMelt'].flatten() / 10 # cm to mm
melt = pd.DataFrame(np.array([X,Y,m,e]).T, columns=['x','y','melt','err']).dropna()
melt.to_csv('./tmp/melt.csv', header=False, index=False)
melt.head()
| x | y | melt | err | |
|---|---|---|---|---|
| 148741 | 1.045e+06 | -2.14e+06 | 1e-09 | 1.71243e-25 |
| 149859 | 1.03e+06 | -2.135e+06 | 0.00146608 | 0.000148305 |
| 149860 | 1.035e+06 | -2.135e+06 | 0.000266042 | 0.000389444 |
| 149861 | 1.04e+06 | -2.135e+06 | 1e-09 | 1.71243e-25 |
| 149862 | 1.045e+06 | -2.135e+06 | 0.00045698 | 0.000668948 |
grass ./G_AQ/PERMANENT
g.mapset -c liefferinge_2023
r.in.xyz input=./tmp/melt.csv output=melt sep=, --o
r.in.xyz input=./tmp/melt.csv output=err z=4 sep=, --oecho "All: " $(r.univar -g map=melt | grep sum)
echo "All: " $(r.univar -g map=err | grep sum)
echo ""
r.univar -gt map=melt zones=basins | cut -d"|" -f2,13 | column -s"|" -t
echo ""
r.univar -gt map=err zones=basins | cut -d"|" -f2,13 | column -s"|" -tAll: sum=69.3982306335468 All: sum=20.0261054475124 label sum East 46.7540492694752 West 18.8528624157926 Peninsula 3.18704264192471 label sum East 14.784924204035 West 4.90322548927998 Peninsula 0.221183549670513
20/69 % = 28.9855072464
- Davison (2023) https://doi.org/10.1126/sciadv.adi0186
import pandas as pd
fname = '~/data/Davison_2023/adi0186_table_s2.xlsx'
loc = pd.read_excel(fname, sheet_name='Total mass changes', index_col = 0, usecols = 'B,C,D', skiprows = 4)
loc = loc.drop('Antarctic Ice Shelves')
df = pd.read_excel(fname, sheet_name='Steady-state',
index_col = 0, skiprows = 4, usecols=((1,4)))
df.columns = ['Mass']
df = loc.join(df)
import geopandas as gpd
fname = '~/data/NSIDC/NSIDC-0709.002/1992.02.07/IceBoundaries_Antarctica_v02.shp'
ew = gpd.read_file(fname)
df = gpd.GeoDataFrame(df, geometry=gpd.points_from_xy(df['longitude'],df['latitude']), crs="EPSG:4326")
df = df.to_crs('epsg:3031')
e = ew.to_crs('epsg:3031')
idx = ew.sindex.nearest(df['geometry'], return_all=False)
df['Region'] = ''
for dfidx,ewidx in idx.T:
arr = df.iloc[dfidx].copy(deep=True)
arr['Region'] = ew.iloc[ewidx]['Regions']
df.iloc[dfidx] = arr
df = df.drop(columns=['latitude','longitude'])
df.loc['Total'] = [df['Mass'].sum(), None, 'All']
df[['Mass','Region']].groupby('Region').sum().drop('Islands').round()
/tmp/ipykernel_3346806/3471234904.py:32: FutureWarning: The behavior of DataFrame concatenation with empty or all-NA entries is deprecated. In a future version, this will no longer exclude empty or all-NA columns when determining the result dtypes. To retain the old behavior, exclude the relevant entries before the concat operation. df.loc['Total'] = [df['Mass'].sum(), None, 'All']
| Region | Mass |
|---|---|
| All | 902.775 |
| East | 392.012 |
| Peninsula | 101.994 |
| West | 408.457 |
Same as above, different sheet. Reuses variables from above, run that first.
fname = '~/data/Davison_2023/adi0186_table_s2.xlsx'
df = pd.read_excel(fname, sheet_name='Steady-state',
index_col = 0, skiprows = 4, usecols=((1,6)))
df.columns = ['Mass']
df = loc.join(df)
df = gpd.GeoDataFrame(df, geometry=gpd.points_from_xy(df['longitude'],df['latitude']), crs="EPSG:4326")
df = df.to_crs('epsg:3031')
e = ew.to_crs('epsg:3031')
idx = ew.sindex.nearest(df['geometry'], return_all=False)
df['Region'] = ''
for dfidx,ewidx in idx.T:
arr = df.iloc[dfidx].copy(deep=True)
arr['Region'] = ew.iloc[ewidx]['Regions']
df.iloc[dfidx] = arr
df = df.drop(columns=['latitude','longitude'])
df.loc['Total'] = [df['Mass'].sum(), None, 'All']
df[['Mass','Region']].groupby('Region').sum().drop('Islands').round()
/tmp/ipykernel_3346806/353247760.py:22: FutureWarning: The behavior of DataFrame concatenation with empty or all-NA entries is deprecated. In a future version, this will no longer exclude empty or all-NA columns when determining the result dtypes. To retain the old behavior, exclude the relevant entries before the concat operation. df.loc['Total'] = [df['Mass'].sum(), None, 'All']
| Region | Mass |
|---|---|
| All | 1348.02 |
| East | 681.734 |
| Peninsula | 103.439 |
| West | 561.832 |
Same as above, different sheet. Reuses variables from above, run that first.
fname = '~/data/Davison_2023/adi0186_table_s2.xlsx'
df = pd.read_excel(fname, sheet_name='Steady-state',
index_col = 0, skiprows = 4, usecols=((1,2)))
df.columns = ['Mass']
df = loc.join(df)
df = gpd.GeoDataFrame(df, geometry=gpd.points_from_xy(df['longitude'],df['latitude']), crs="EPSG:4326")
df = df.to_crs('epsg:3031')
e = ew.to_crs('epsg:3031')
idx = ew.sindex.nearest(df['geometry'], return_all=False)
df['Region'] = ''
for dfidx,ewidx in idx.T:
arr = df.iloc[dfidx].copy(deep=True)
arr['Region'] = ew.iloc[ewidx]['Regions']
df.iloc[dfidx] = arr
df = df.drop(columns=['latitude','longitude'])
df.loc['Total'] = [df['Mass'].sum(), None, 'All']
df[['Mass','Region']].groupby('Region').sum().drop('Islands').round()
/tmp/ipykernel_3346806/927385710.py:22: FutureWarning: The behavior of DataFrame concatenation with empty or all-NA entries is deprecated. In a future version, this will no longer exclude empty or all-NA columns when determining the result dtypes. To retain the old behavior, exclude the relevant entries before the concat operation. df.loc['Total'] = [df['Mass'].sum(), None, 'All']
| Region | Mass |
|---|---|
| All | 1838.8 |
| East | 910.573 |
| Peninsula | 159.697 |
| West | 767.324 |
[greene_Supplementary_Table_1.xlsx](https://github.com/user-attachments/files/15598602/greene_Supplementary_Table_1.xlsx)
I think the data in the attached spreadsheet from [Greene et al., 2022 ](https://doi.org/10.1038/s41586-022-05037-w) is everything needed for ice-shelf mass-change resulting from frontal advance/retreat, so in Excel `=BI189-O189` gives Antarctica’s net retreat from 1997 to 2021. Change the column to adjust the time period.
BI189 = 24596304.0 BI189 = 2021.2 Q189 = 24597630.0 Q189 = 2000.2
(24596304.0 - 24597630.0) / (2021.2-2000.2) = -63.1428571429
But we need to recreate this in code so we can split by east/west/peninsula
import pandas as pd
import geopandas as gpd
fname = "~/data/Greene_2022/data/greene_Supplementary_Table_1.xlsx"
df = pd.read_excel(fname, sheet_name='greene_iceshelf_area_and_mass',
index_col = 1, skiprows = 4)
df = df.rename(columns={'Unnamed: 2':'lat',
'Unnamed: 3':'lon'})
# drop uncertainty columns
unc = []
for c in df.columns:
if type(c) == str:
if c[0:8] == 'Unnamed:':
unc.append(c)
df = df.drop(columns = unc)
df = df[['lat','lon',2000.2,2021.2]]
df = df.iloc[1:]
# Remove last two rows
aq = df.loc['Antarctica']
other = df.loc['Other']
df = df.iloc[:-2]
print(df.sum())
print("")
print(aq)
print("")
print(other)
lat -12882.373098 lon 6279.268331 2000.2 682491.281291 2021.2 681213.775349 dtype: object lat -90 lon every 2000.2 24597630 2021.2 24596304 Name: Antarctica, dtype: object lat NaN lon NaN 2000.2 23915136 2021.2 23915090 Name: Other, dtype: object
shelf = df.sum()
print("All AQ loss: ", (aq[2021.2] - aq[2000.2]) / (2021-2000))
print("Named shelf loss: ", (shelf[2021.2] - shelf[2000.2]) / (2021-2000))
print("Other loss: ", (other[2021.2] - other[2000.2]) / (2021-2000))
print("Named + Other: ", (((other + shelf)[2021.2] - (other + shelf)[2000.2]) / (2021-2000)))
print("Named %: ", 2.19/63.02*100)
All AQ loss: -63.142857142857146 Named shelf loss: -60.83361628651619 Other loss: -2.1904761904761907 Named + Other: -63.02409247699238 Named %: 3.4750872738813077
import geopandas as gpd
fname = '~/data/NSIDC/NSIDC-0709.002/1992.02.07/IceBoundaries_Antarctica_v02.shp'
ew = gpd.read_file(fname)
ew.drop(columns=['geometry']).head()
| NAME | Regions | Subregions | TYPE | Asso_Shelf | |
|---|---|---|---|---|---|
| 0 | LarsenE | Peninsula | Ipp-J | GR | LarsenE |
| 1 | Dawson_Lambton | East | nan | FL | nan |
| 2 | Academy | East | Jpp-K | GR | Filchner |
| 3 | Brunt_Stancomb | East | K-A | GR | Brunt_Stancomb |
| 4 | Riiser-Larsen | East | K-A | GR | Riiser-Larsen |
gdf = gpd.GeoDataFrame(df, geometry=gpd.points_from_xy(df['lon'],df['lat']), crs="EPSG:4326")
gdf = gdf.to_crs('epsg:3031')
ew = ew.to_crs('epsg:3031')
idx = ew.sindex.nearest(gdf['geometry'], return_all=False)
gdf['Region'] = ''
for gdfidx,ewidx in idx.T:
arr = gdf.iloc[gdfidx].copy(deep=True)
arr['Region'] = ew.iloc[ewidx]['Regions']
gdf.iloc[gdfidx] = arr
gdf.head()
gdf.loc['Total'] = gdf.sum(axis='rows')
gdf.loc['Total', 'Region'] = 'All'
gdf['frontal change'] = (gdf[2021.2] - gdf[2000.2]) / (2021.2-2000.2)
pos = gdf[gdf['frontal change'] > 0]
neg = gdf[gdf['frontal change'] <= 0]
# gdf
print('neg', neg[['Region','frontal change']].groupby('Region').sum().round().abs())
print('')
print('pos', pos[['Region','frontal change']].groupby('Region').sum().round().abs())
print('')
print('all', gdf[['Region','frontal change']].groupby('Region').sum().round().abs())
neg frontal change Region All 61.0 East 79.0 Peninsula 122.0 West 145.0 pos frontal change Region East 181.0 Peninsula 1.0 West 103.0 all frontal change Region All 61.0 East 102.0 Peninsula 121.0 West 42.0
Email from Davison
| Ice Shelf | Mass change due to grounding line migration from 1997 to 2021 (Gt) | Error (Gt) |
| Pine Island | 220 | 40 |
| Thwaites | 230 | 25 |
| Crosson | 200 | 25 |
| Dotson | 420 | 80 |
(220+230+200+420)/(2021-1997) = 44.5833333333
import xarray as xr
ds = xr.open_mfdataset("~/data/Paolo_2023/ANT_G1920V01_IceShelfMelt.nc")
ds = ds[['melt','melt_err']].sel({'time':slice('2000-01-01','2017-12-31')}).mean(dim='time')
delayed_obj = ds.to_netcdf('tmp/shelf_melt.nc', compute=False)
from dask.diagnostics import ProgressBar
with ProgressBar():
results = delayed_obj.compute()
print(ds)
[########################################] | 100% Completed | 7.79 s
<xarray.Dataset> Size: 68MB
Dimensions: (y: 2916, x: 2916)
Coordinates:
* x (x) float64 23kB -2.798e+06 -2.796e+06 ... 2.796e+06 2.798e+06
* y (y) float64 23kB 2.798e+06 2.796e+06 ... -2.796e+06 -2.798e+06
Data variables:
melt (y, x) float32 34MB dask.array<chunksize=(486, 486), meta=np.ndarray>
melt_err (y, x) float32 34MB dask.array<chunksize=(486, 486), meta=np.ndarray>
g.mapset -c Paolo_2023
ncdump -v x tmp/shelf_melt.nc # 2916x2916
ncdump -v y tmp/shelf_melt.nc
x0=-2798407.5
x1=2798392.5
y0=-2798392.5
y1=2798407.5
g.region w=$(( -2798407 - 960 )) e=$(( 2798392 + 960 )) s=$(( -2798392 - 960 )) n=$(( 2798407 + 960 )) res=1920 -p
r.mapcalc "area = area()"
r.in.gdal -o input=NetCDF:tmp/shelf_melt.nc:melt output=melt
r.in.gdal -o input=NetCDF:tmp/shelf_melt.nc:melt_err output=err
r.region -c map=melt
r.region -c map=err
## + kg/m^2 -> Gt: / 1E12
r.mapcalc "melt = melt * 1000 * area / exp(10,12)" --o
r.mapcalc "err = err * 1000 * area / exp(10,12)" --o
r.mapcalc "melt_on = if(melt > 0, melt, null())"
r.mapcalc "err_on = if(melt > 0, err, null())"
r.mapcalc "melt_off = if(melt < 0, melt, null())"
r.mapcalc "err_off = if(melt < 0, err, null())"
r.colors -ae map=melt color=difference
r.colors -ge map=melt_on color=viridis
r.colors -ge map=melt_off color=viridis
# d.rast melt
# d.rast melt_on
# d.rast melt_off
r.mapcalc "basins = if((basins@PERMANENT == 1) | (basins@PERMANENT == 11), 1, 0)"
r.mapcalc "basins = if((basins@PERMANENT == 2) | (basins@PERMANENT == 12), 2, basins)"
r.mapcalc "basins = if((basins@PERMANENT == 3) | (basins@PERMANENT == 13), 3, basins)"
r.colors map=basins color=viridis
r.category basins separator=":" rules=- << EOF
1:East
2:West
3:Peninsula
EOFecho "NET"
r.univar -gt map=melt zones=basins | cut -d"|" -f2,13 | column -s"|" -t | sed 's/label.*//'
r.univar -gt map=err zones=basins | cut -d"|" -f2,13 | column -s"|" -t | sed 's/label.*//'
# r.univar -g melt | grep sum
echo ""
echo "FREEZE_ON"
r.univar -gt map=melt_on zones=basins | cut -d"|" -f2,13 | column -s"|" -t | sed 's/label.*//'
r.univar -gt map=err_on zones=basins | cut -d"|" -f2,13 | column -s"|" -t | sed 's/label.*//'
# r.univar -g melt_on | grep sum
echo ""
echo "MELT_OFF"
r.univar -gt map=melt_off zones=basins | cut -d"|" -f2,13 | column -s"|" -t | sed 's/label.*//'
r.univar -gt map=err_off zones=basins | cut -d"|" -f2,13 | column -s"|" -t | sed 's/label.*//'
# r.univar -g melt_off | grep sumNET East -314.59815832386 West -516.40649095369 Peninsula -144.781596164836 East 1262.04130311163 West 1500.90827154852 Peninsula 230.826477084729 FREEZE_ON East 199.82814766386 West 146.110647983701 Peninsula 10.5669662518996 East 462.492239471166 West 576.342637546049 Peninsula 35.4516506207232 MELT_OFF East -514.426305987718 West -662.517138937384 Peninsula -155.348562416735 East 799.549063640453 West 924.565634002478 Peninsula 195.374826464004
(save-excursion
(goto-char (point-min))
(re-search-forward (concat "^#\\+name: " tbl) nil t)
(next-line)
(org-table-export (concat "./dat/" tbl ".csv") "orgtbl-to-csv")
;;(shell-command-to-string (concat "head " tbl ".csv"))
(message (concat "Exported " tbl " to " (concat "./dat/" tbl ".csv")))
)