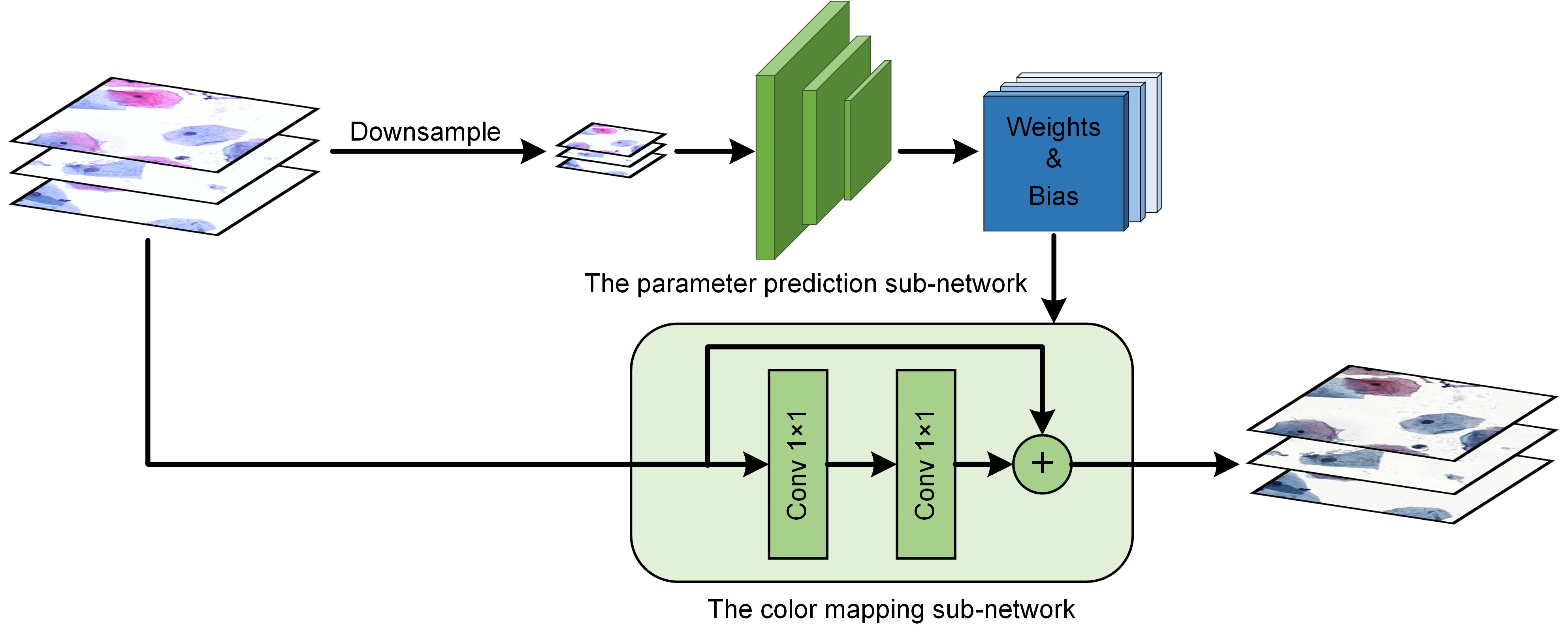
import numpy as np
import torch
from source .model import ParamNet
from PIL import Image
import matplotlib .pyplot as plt
% matplotlib inline def norm (image ):
image = np .array (image ).astype (np .float32 )
image = image .transpose ((2 , 0 , 1 ))
image = ((image / 255 ) - 0.5 ) / 0.5
image = image [np .newaxis , ...]
image = torch .from_numpy (image )
return image
def un_norm (image ):
image = image .cpu ().detach ().numpy ()[0 ]
image = ((image * 0.5 + 0.5 ) * 255 ).astype (np .uint8 ).transpose ((1 ,2 ,0 ))
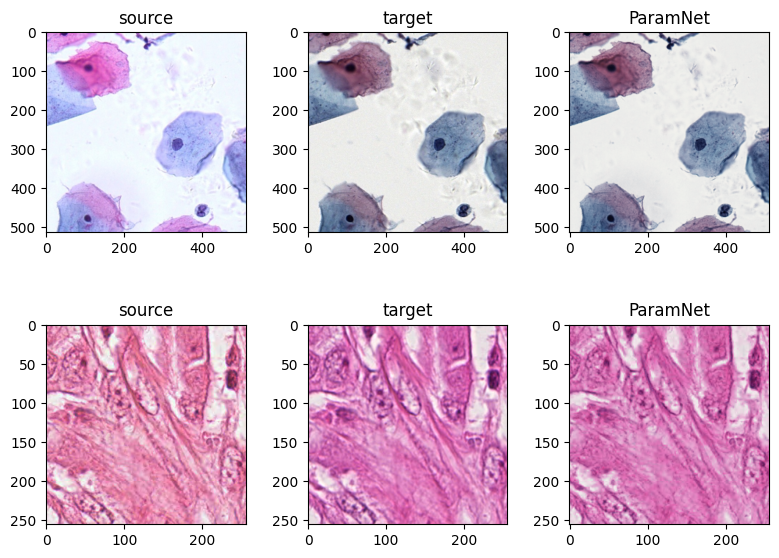
return image The demo for one-to-one stain normalization #read source image
img_source_cyp = Image .open ("assets/aligned/aligned_cyp_source.png" )
img_target_cyp = Image .open ("assets/aligned/aligned_cyp_target.png" )
img_source_hist = Image .open ("assets/aligned/aligned_hist_source.png" )
img_target_hist = Image .open ("assets/aligned/aligned_hist_target.png" )# run on the aligned dataset
model_cyp = ParamNet ()
model_cyp .load_state_dict (torch .load ("checkpoints/ParamNet-source.pt" )['net_G_A' ])
img_norm_cyp = model_cyp (norm (img_source_cyp ))
img_norm_cyp = un_norm (img_norm_cyp )
model_hist = ParamNet ()
model_hist .load_state_dict (torch .load ("checkpoints/ParamNet-aperio.pt" )['net_G_A' ])
img_norm_hist = model_hist (norm (img_source_hist ))
img_norm_hist = un_norm (img_norm_hist )fig ,axes = plt .subplots (2 ,3 ,figsize = (8 , 6 ))
#plt.subplots_adjust(wspace =0.25, hspace =0.25)#调整子图间距
fig .tight_layout ()
axes [0 ,0 ].imshow (img_source_cyp )
axes [0 ,0 ].set_title ('source' )
axes [0 ,1 ].imshow (img_target_cyp )
axes [0 ,1 ].set_title ('target' )
axes [0 ,2 ].imshow (img_norm_cyp )
axes [0 ,2 ].set_title ('ParamNet' )
axes [1 ,0 ].imshow (img_source_hist )
axes [1 ,0 ].set_title ('source' )
axes [1 ,1 ].imshow (img_target_hist )
axes [1 ,1 ].set_title ('target' )
axes [1 ,2 ].imshow (img_norm_hist )
axes [1 ,2 ].set_title ('ParamNet' )
plt .show ()
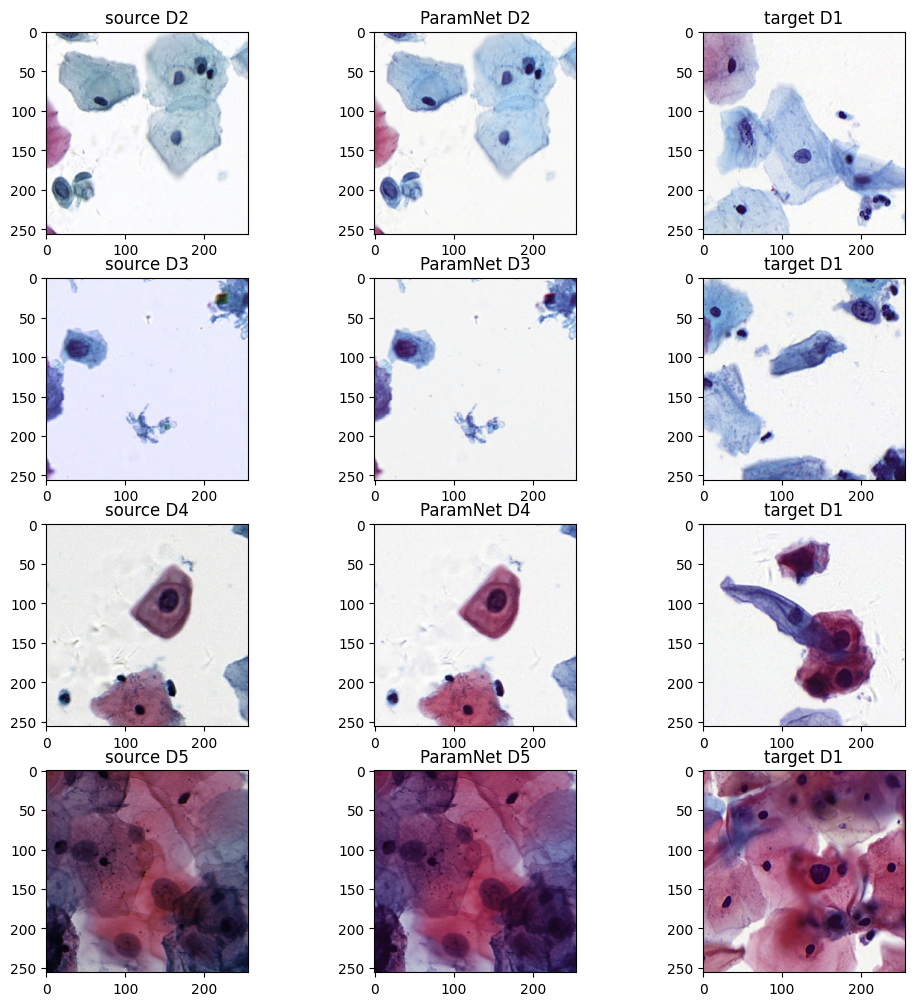
The demo for multi-to-one stain normalization 1. Cyptopathology Dataset #read source image
img_D2 = Image .open ("assets/D2-D5/ori_D2.png" )
img_D3 = Image .open ("assets/D2-D5/ori_D3.png" )
img_D4 = Image .open ("assets/D2-D5/ori_D4.png" )
img_D5 = Image .open ("assets/D2-D5/ori_D5.png" )
img_D1_1 = Image .open ("assets/D1/1.png" )
img_D1_2 = Image .open ("assets/D1/2.png" )
img_D1_3 = Image .open ("assets/D1/3.png" )
img_D1_4 = Image .open ("assets/D1/4.png" )model_D2_5 = ParamNet ()
model_D2_5 .load_state_dict (torch .load ("checkpoints/ParamNet-D1.pt" )['net_G_A' ])
img_norm_D2 = model_D2_5 (norm (img_D2 ))
img_norm_D2 = un_norm (img_norm_D2 )
img_norm_D3 = model_D2_5 (norm (img_D3 ))
img_norm_D3 = un_norm (img_norm_D3 )
img_norm_D4 = model_D2_5 (norm (img_D4 ))
img_norm_D4 = un_norm (img_norm_D4 )
img_norm_D5 = model_D2_5 (norm (img_D5 ))
img_norm_D5 = un_norm (img_norm_D5 )fig ,axes = plt .subplots (4 ,3 ,figsize = (10 , 10 ))
#plt.subplots_adjust(wspace =0.25, hspace =0.25)#调整子图间距
fig .tight_layout ()
axes [0 ,0 ].imshow (img_D2 )
axes [0 ,0 ].set_title ('source D2' )
axes [1 ,0 ].imshow (img_D3 )
axes [1 ,0 ].set_title ('source D3' )
axes [2 ,0 ].imshow (img_D4 )
axes [2 ,0 ].set_title ('source D4' )
axes [3 ,0 ].imshow (img_D5 )
axes [3 ,0 ].set_title ('source D5' )
axes [0 ,1 ].imshow (img_norm_D2 )
axes [0 ,1 ].set_title ('ParamNet D2' )
axes [1 ,1 ].imshow (img_norm_D3 )
axes [1 ,1 ].set_title ('ParamNet D3' )
axes [2 ,1 ].imshow (img_norm_D4 )
axes [2 ,1 ].set_title ('ParamNet D4' )
axes [3 ,1 ].imshow (img_norm_D5 )
axes [3 ,1 ].set_title ('ParamNet D5' )
axes [0 ,2 ].imshow (img_D1_1 )
axes [0 ,2 ].set_title ('target D1' )
axes [1 ,2 ].imshow (img_D1_2 )
axes [1 ,2 ].set_title ('target D1' )
axes [2 ,2 ].imshow (img_D1_3 )
axes [2 ,2 ].set_title ('target D1' )
axes [3 ,2 ].imshow (img_D1_4 )
axes [3 ,2 ].set_title ('target D1' )
plt .show ()
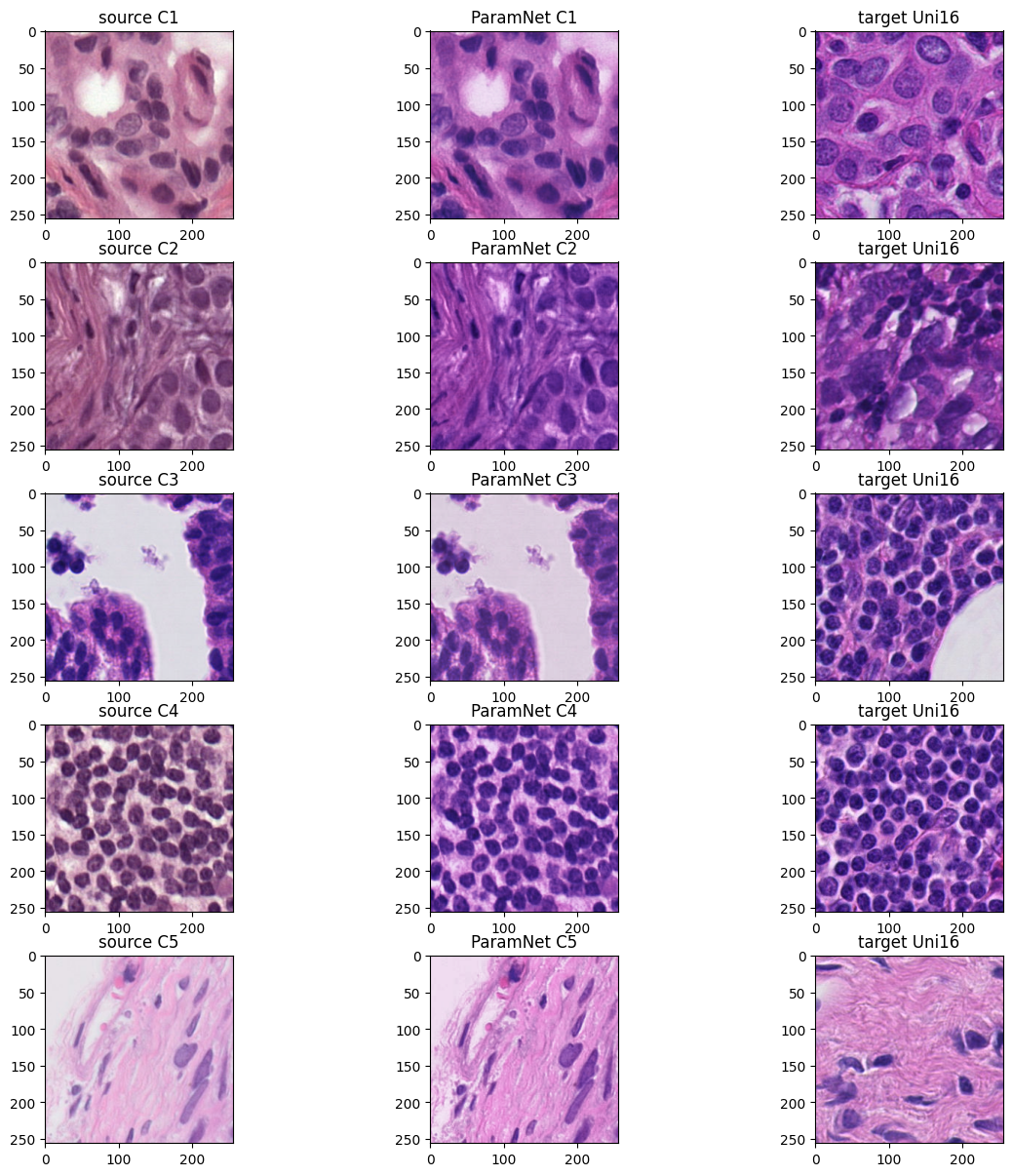
2. Histopathology Dataset #read source image
img_C1 = Image .open ("assets/C1-C5/source_C1.png" )
img_C2 = Image .open ("assets/C1-C5/source_C2.png" )
img_C3 = Image .open ("assets/C1-C5/source_C3.png" )
img_C4 = Image .open ("assets/C1-C5/source_C4.png" )
img_C5 = Image .open ("assets/C1-C5/source_C5.png" )
img_Uni16_1 = Image .open ("assets/Uni16/Uni_1.png" )
img_Uni16_2 = Image .open ("assets/Uni16/Uni_2.png" )
img_Uni16_3 = Image .open ("assets/Uni16/Uni_3.png" )
img_Uni16_4 = Image .open ("assets/Uni16/Uni_4.png" )
img_Uni16_5 = Image .open ("assets/Uni16/Uni_5.png" )model_C1_5 = ParamNet ()
model_C1_5 .load_state_dict (torch .load ("checkpoints/ParamNet-Uni.pt" )['net_G_A' ])
img_norm_C1 = model_C1_5 (norm (img_C1 ))
img_norm_C1 = un_norm (img_norm_C1 )
img_norm_C2 = model_C1_5 (norm (img_C2 ))
img_norm_C2 = un_norm (img_norm_C2 )
img_norm_C3 = model_C1_5 (norm (img_C3 ))
img_norm_C3 = un_norm (img_norm_C3 )
img_norm_C4 = model_C1_5 (norm (img_C4 ))
img_norm_C4 = un_norm (img_norm_C4 )
img_norm_C5 = model_C1_5 (norm (img_C5 ))
img_norm_C5 = un_norm (img_norm_C5 )
fig ,axes = plt .subplots (5 ,3 ,figsize = (12 , 12 ))
#plt.subplots_adjust(wspace =0.25, hspace =0.25)#调整子图间距
fig .tight_layout ()
axes [0 ,0 ].imshow (img_C1 )
axes [0 ,0 ].set_title ('source C1' )
axes [1 ,0 ].imshow (img_C2 )
axes [1 ,0 ].set_title ('source C2' )
axes [2 ,0 ].imshow (img_C3 )
axes [2 ,0 ].set_title ('source C3' )
axes [3 ,0 ].imshow (img_C4 )
axes [3 ,0 ].set_title ('source C4' )
axes [4 ,0 ].imshow (img_C5 )
axes [4 ,0 ].set_title ('source C5' )
axes [0 ,1 ].imshow (img_norm_C1 )
axes [0 ,1 ].set_title ('ParamNet C1' )
axes [1 ,1 ].imshow (img_norm_C2 )
axes [1 ,1 ].set_title ('ParamNet C2' )
axes [2 ,1 ].imshow (img_norm_C3 )
axes [2 ,1 ].set_title ('ParamNet C3' )
axes [3 ,1 ].imshow (img_norm_C4 )
axes [3 ,1 ].set_title ('ParamNet C4' )
axes [4 ,1 ].imshow (img_norm_C5 )
axes [4 ,1 ].set_title ('ParamNet C5' )
axes [0 ,2 ].imshow (img_Uni16_1 )
axes [0 ,2 ].set_title ('target Uni16' )
axes [1 ,2 ].imshow (img_Uni16_2 )
axes [1 ,2 ].set_title ('target Uni16' )
axes [2 ,2 ].imshow (img_Uni16_3 )
axes [2 ,2 ].set_title ('target Uni16' )
axes [3 ,2 ].imshow (img_Uni16_4 )
axes [3 ,2 ].set_title ('target Uni16' )
axes [4 ,2 ].imshow (img_Uni16_5 )
axes [4 ,2 ].set_title ('target Uni16' )
plt .show ()